Secondary Sugar Binding Site Identified for Leca Lectin from Pseudomonas Aeruginosa.
Blanchard, B., Imberty, A., Varrot, A.(2014) Proteins 82: 1060
- PubMed: 24123124
- DOI: https://doi.org/10.1002/prot.24430
- Primary Citation of Related Structures:
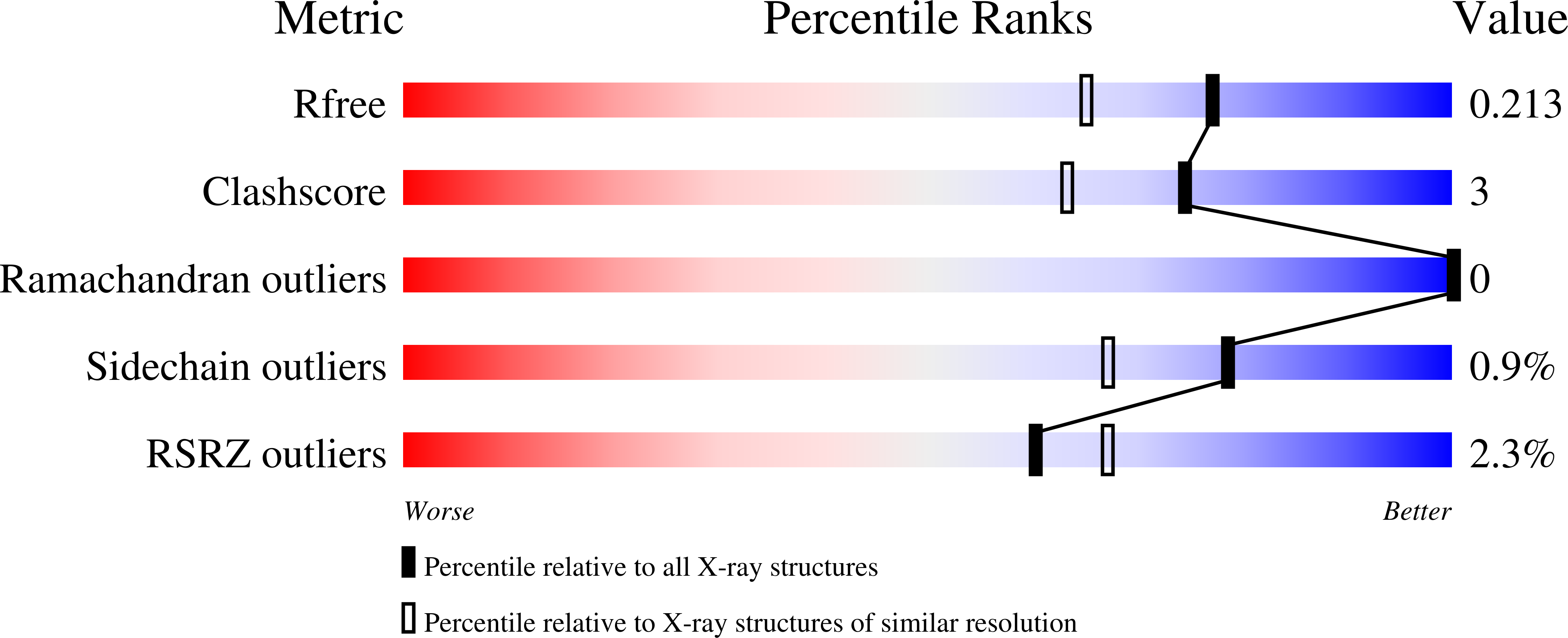
4AL9 - PubMed Abstract:
The galactose-specific lectin LecA from Pseudomonas aeruginosa is a target for the development of new anti-infectious compounds. Sugar based molecules with anti-adhesive properties present great potential in the fight against bacterial infection and biofilm formation. LecA is specific for oligosaccharides with terminal α-galactoside residues and displays strong affinity for melibiose (αGal1-6Glc) with a Kd of 38.8 µM. The crystal structure of LecA/melibiose complex shows classical calcium-bridged binding of αGal in the primary binding site but also revealed a secondary sugar binding site with glucose bound. This sugar binding site is in close proximity to the galactose binding one, is independent of calcium and mainly involves interactions with a symmetry-related protein. This discovery would help to the design of new potent inhibitors targeting both binding sites.
Organizational Affiliation:
CERMAV-CNRS, Université de Grenoble and Member of ICMG, BP 53, 38041 Grenoble Cedex 9, France.