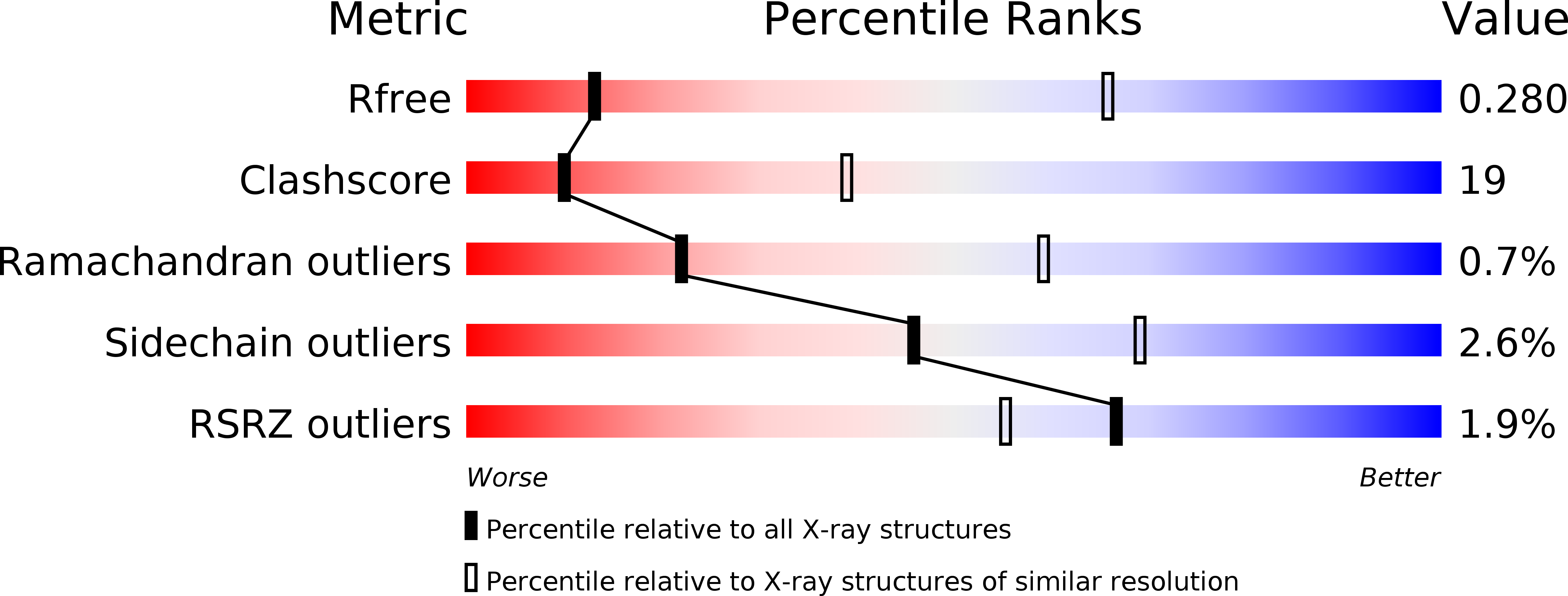
Thermodynamic Properties Distinguish Human Mitochondrial Aspartyl-tRNA Synthetase from Bacterial Homolog with Same 3D Architecture
Ennifar, E., Florentz, C., Gaudry, A., Lorber, B., Neuenfeldt, A., Sauter, C., Sissler, M.(2013) Nucleic Acids Res 41: 2698
- PubMed: 23275545
- DOI: https://doi.org/10.1093/nar/gks1322
- Primary Citation of Related Structures:
4AH6 - PubMed Abstract:
In the mammalian mitochondrial translation apparatus, the proteins and their partner RNAs are coded by two genomes. The proteins are nuclear-encoded and resemble their homologs, whereas the RNAs coming from the rapidly evolving mitochondrial genome have lost critical structural information. This raises the question of molecular adaptation of these proteins to their peculiar partner RNAs. The crystal structure of the homodimeric bacterial-type human mitochondrial aspartyl-tRNA synthetase (DRS) confirmed a 3D architecture close to that of Escherichia coli DRS. However, the mitochondrial enzyme distinguishes by an enlarged catalytic groove, a more electropositive surface potential and an alternate interaction network at the subunits interface. It also presented a thermal stability reduced by as much as 12°C. Isothermal titration calorimetry analyses revealed that the affinity of the mitochondrial enzyme for cognate and non-cognate tRNAs is one order of magnitude higher, but with different enthalpy and entropy contributions. They further indicated that both enzymes bind an adenylate analog by a cooperative allosteric mechanism with different thermodynamic contributions. The larger flexibility of the mitochondrial synthetase with respect to the bacterial enzyme, in combination with a preserved architecture, may represent an evolutionary process, allowing nuclear-encoded proteins to cooperate with degenerated organelle RNAs.
Organizational Affiliation:
Architecture et Réactivité de l'ARN, Université de Strasbourg, CNRS, IBMC, F-67084 Strasbourg Cedex, France.