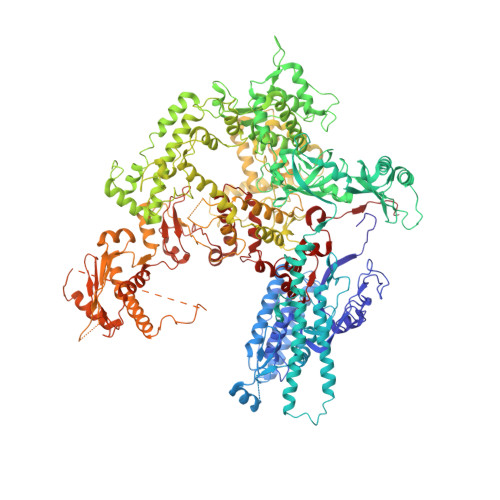
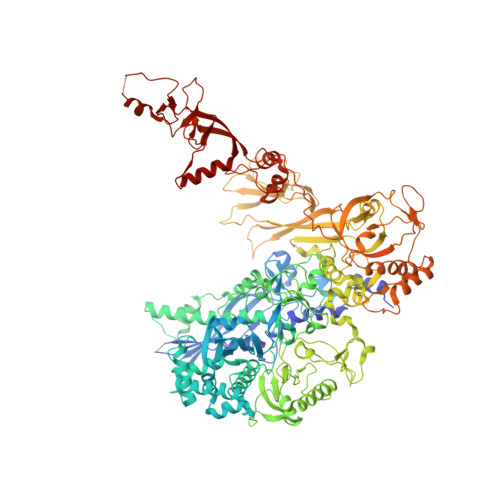
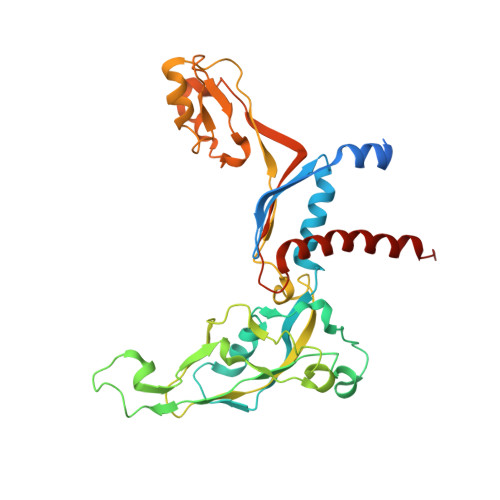
An alternative RNA polymerase I structure reveals a dimer hinge.
Kostrewa, D., Kuhn, C.D., Engel, C., Cramer, P.(2015) Acta Crystallogr D Biol Crystallogr 71: 1850-1855
- PubMed: 26327374
- DOI: https://doi.org/10.1107/S1399004715012651
- Primary Citation of Related Structures:
4YM7 - PubMed Abstract:
RNA polymerase I (Pol I) is the central, 14-subunit enzyme that synthesizes the ribosomal RNA (rRNA) precursor in eukaryotic cells. The recent crystal structure of Pol I at 2.8 Å resolution revealed two novel elements: the `expander' in the active-centre cleft and the `connector' that mediates Pol I dimerization [Engel et al. (2013), Nature (London), 502, 650-655]. Here, a Pol I structure in an alternative crystal form that was solved by molecular replacement using the original atomic Pol I structure is reported. The resulting alternative structure lacks the expander but still shows an expanded active-centre cleft. The neighbouring Pol I monomers form a homodimer with a relative orientation distinct from that observed previously, establishing the connector as a hinge between Pol I monomers.
Organizational Affiliation:
Gene Center Munich and Department of Biochemistry, Ludwig-Maximilians-Universität München, Feodor-Lynen-Strasse 25, 81377 Munich, Germany.