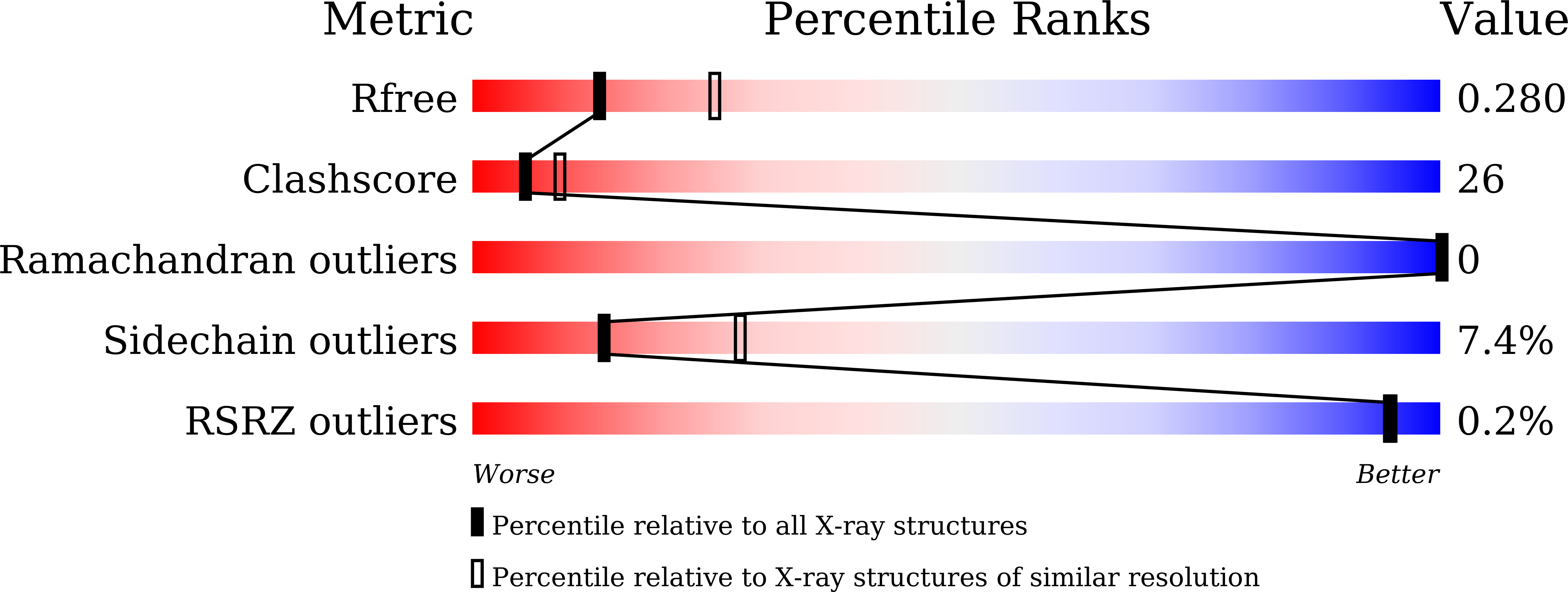
A secreted protein with plant-specific cysteine-rich motif functions as a mannose-binding lectin that exhibits antifungal activity.
Miyakawa, T., Hatano, K., Miyauchi, Y., Suwa, Y., Sawano, Y., Tanokura, M.(2014) Plant Physiol 166: 766-778
- PubMed: 25139159
- DOI: https://doi.org/10.1104/pp.114.242636
- Primary Citation of Related Structures:
4XRE - PubMed Abstract:
Plants have a variety of mechanisms for defending against plant pathogens and tolerating environmental stresses such as drought and high salinity. Ginkbilobin2 (Gnk2) is a seed storage protein in gymnosperm that possesses antifungal activity and a plant-specific cysteine-rich motif (domain of unknown function26 [DUF26]). The Gnk2-homologous sequence is also observed in an extracellular region of cysteine-rich repeat receptor-like kinases that function in response to biotic and abiotic stresses. Here, we report the lectin-like molecular function of Gnk2 and the structural basis of its monosaccharide recognition. Nuclear magnetic resonance experiments showed that mannan was the only yeast (Saccharomyces cerevisiae) cell wall polysaccharide that interacted with Gnk2. Gnk2 also interacted with mannose, a building block of mannan, with a specificity that was similar to those of mannose-binding legume lectins, by strictly recognizing the configuration of the hydroxy group at the C4 position of the monosaccharide. The crystal structure of Gnk2 in complex with mannose revealed that three residues (asparagine-11, arginine-93, and glutamate-104) recognized mannose by hydrogen bonds, which defined the carbohydrate-binding specificity. These interactions were directly related to the ability of Gnk2 to inhibit the growth of fungi, including the plant pathogenic Fusarium spp., which were disrupted by mutation of arginine-93 or the presence of yeast mannan in the assay system. In addition, Gnk2 did not inhibit the growth of a yeast mutant strain lacking the α1,2-linked mannose moiety. These results provide insights into the molecular basis of the DUF26 protein family.
Organizational Affiliation:
Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, University of Tokyo, Bunkyo-ku, Tokyo 113-8657, Japan (T.M., Y.M., Y.Su., M.T.);Division of Molecular Science, Faculty of Science and Technology, Gunma University, Kiryu, Gunma 376-8515, Japan (K.H.); andLaboratory of Chemistry, College of Liberal Arts and Sciences, Tokyo Medical and Dental University, Ichikawa-shi, Chiba 272-0827, Japan (Y.Sa.).