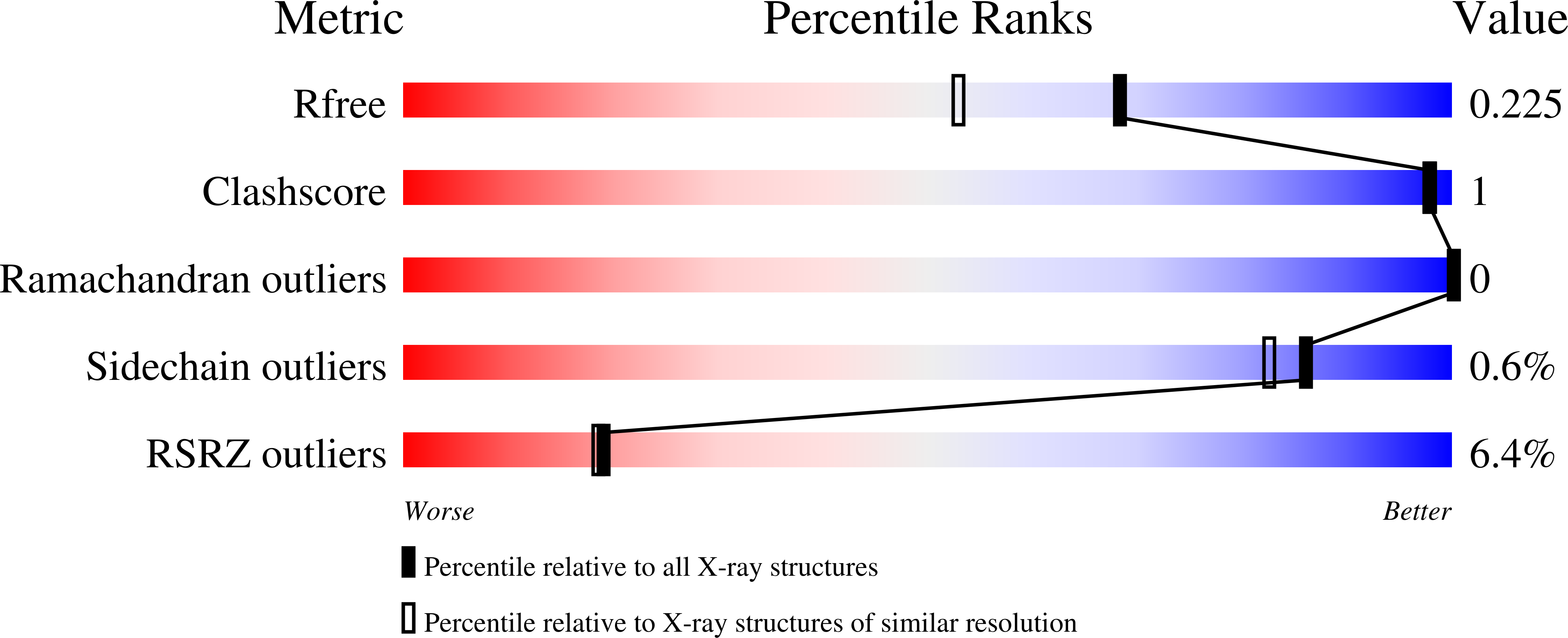
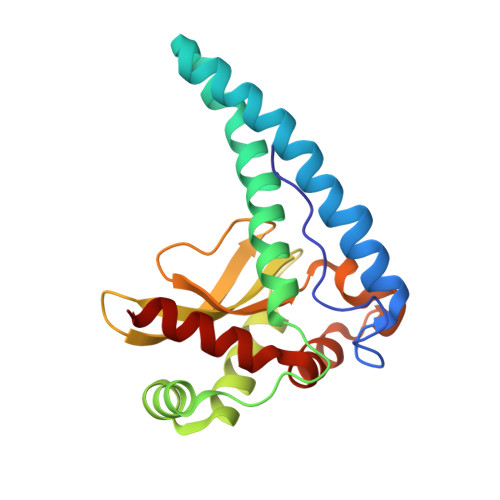
The structure of the Caenorhabditis elegans manganese superoxide dismutase MnSOD-3-azide complex.
Hunter, G.J., Trinh, C.H., Bonetta, R., Stewart, E.E., Cabelli, D.E., Hunter, T.(2015) Protein Sci 24: 1777-1788
- PubMed: 26257399
- DOI: https://doi.org/10.1002/pro.2768
- Primary Citation of Related Structures:
4X9Q, 5AG2 - PubMed Abstract:
C. elegans MnSOD-3 has been implicated in the longevity pathway and its mechanism of catalysis is relevant to the aging process and carcinogenesis. The structures of MnSOD-3 provide unique crystallographic evidence of a dynamic region of the tetrameric interface (residues 41-54). We have determined the structure of the MnSOD-3-azide complex to 1.77-Å resolution. Analysis of this complex shows that the substrate analog, azide, binds end-on to the manganese center as a sixth ligand and that it ligates directly to a third and new solvent molecule also positioned within interacting distance to the His30 and Tyr34 residues of the substrate access funnel. This is the first structure of a eukaryotic MnSOD-azide complex that demonstrates the extended, uninterrupted hydrogen-bonded network that forms a proton relay incorporating three outer sphere solvent molecules, the substrate analog, the gateway residues, Gln142, and the solvent ligand. This configuration supports the formation and release of the hydrogen peroxide product in agreement with the 5-6-5 catalytic mechanism for MnSOD. The high product dissociation constant k4 of MnSOD-3 reflects low product inhibition making this enzyme efficient even at high levels of superoxide.
Organizational Affiliation:
Department of Physiology and Biochemistry, Faculty of Medicine and Surgery, University of Malta, Msida, Malta.