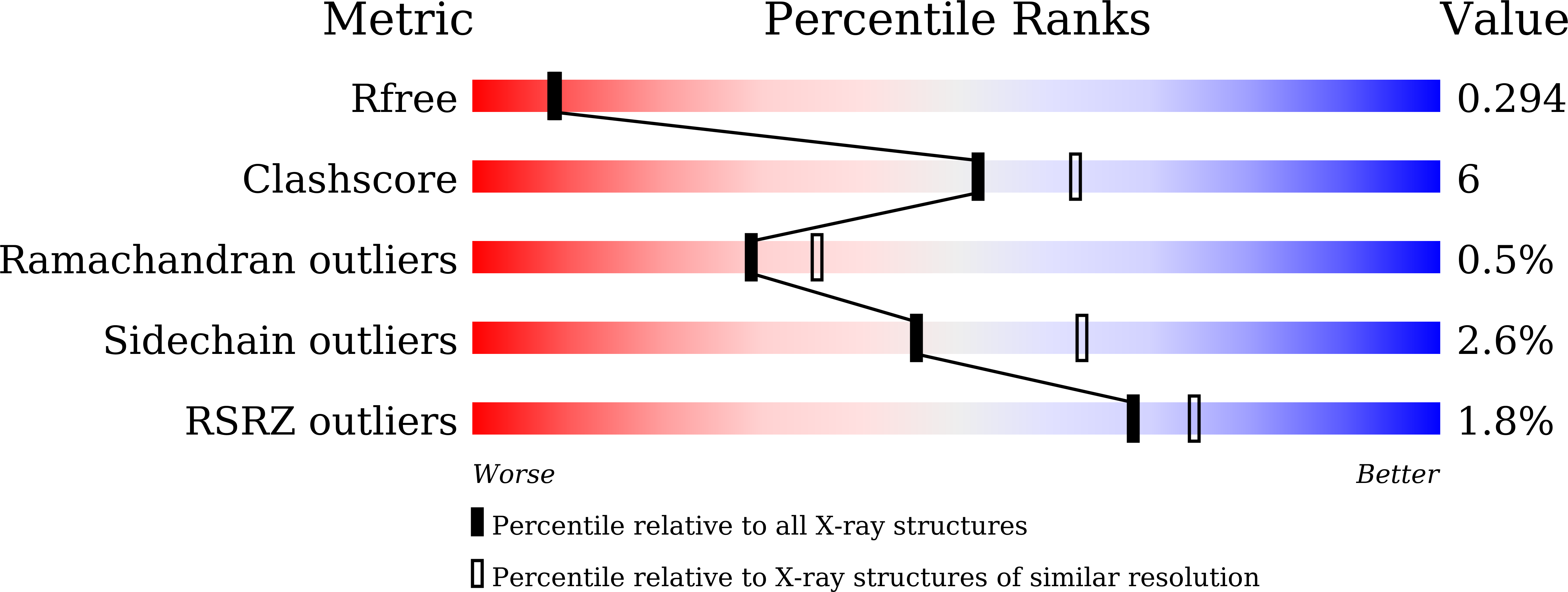
Crystal structure of IMP-2 metallo-beta-lactamase from Acinetobacter spp.: comparison of active-site loop structures between IMP-1 and IMP-2.
Yamaguchi, Y., Matsueda, S., Matsunaga, K., Takashio, N., Toma-Fukai, S., Yamagata, Y., Shibata, N., Wachino, J., Shibayama, K., Arakawa, Y., Kurosaki, H.(2015) Biol Pharm Bull 38: 96-101
- PubMed: 25744464
- DOI: https://doi.org/10.1248/bpb.b14-00594
- Primary Citation of Related Structures:
4UBQ - PubMed Abstract:
IMP-2, a subclass B1 metallo-β-lactamase (MBL), is a Zn(II)-containing hydrolase. This hydrolase, involved in antibiotic resistance, catalyzes the hydrolysis of the C-N bond of the β-lactam ring in β-lactam antibiotics such as benzylpenicillin and imipenem. The crystal structure of IMP-2 MBL from Acinetobacter spp. was determined at 2.3 Å resolution. This structure is analogous to that of subclass B1 MBLs such as IMP-1 and VIM-2. Comparison of the structures of IMP-1 and IMP-2, which have an 85% amino acid identity, suggests that the amino acid substitution at position 68 on a β-strand (β3) (Pro in IMP-1 versus Ser in IMP-2) may be a staple factor affecting the flexibility of loop 1 (comprising residues at positions 60-66; EVNGWGV). In the IMP-1 structure, loop 1 adopts an open, disordered conformation. On the other hand, loop 1 of IMP-2 forms a closed conformation in which the side chain of Trp64, involved in substrate binding, is oriented so as to cover the active site, even though there is an acetate ion in the active site of both IMP-1 and IMP-2. Loop 1 of IMP-2 has a more flexible structure in comparison to IMP-1 due to having a Ser residue instead of the Pro residue at position 68, indicating that this difference in sequence may be a trigger to induce a more flexible conformation in loop 1.
Organizational Affiliation:
Environmental Safety Center, Kumamoto University.