Interfacial residues of SpcS chaperone affects binding of effector toxin ExoT in Pseudomonas aeruginosa: novel insights from structural and computational studies
Dey, S., Datta, S.(2014) FEBS J 281: 1267-1280
- PubMed: 24387107
- DOI: https://doi.org/10.1111/febs.12704
- Primary Citation of Related Structures:
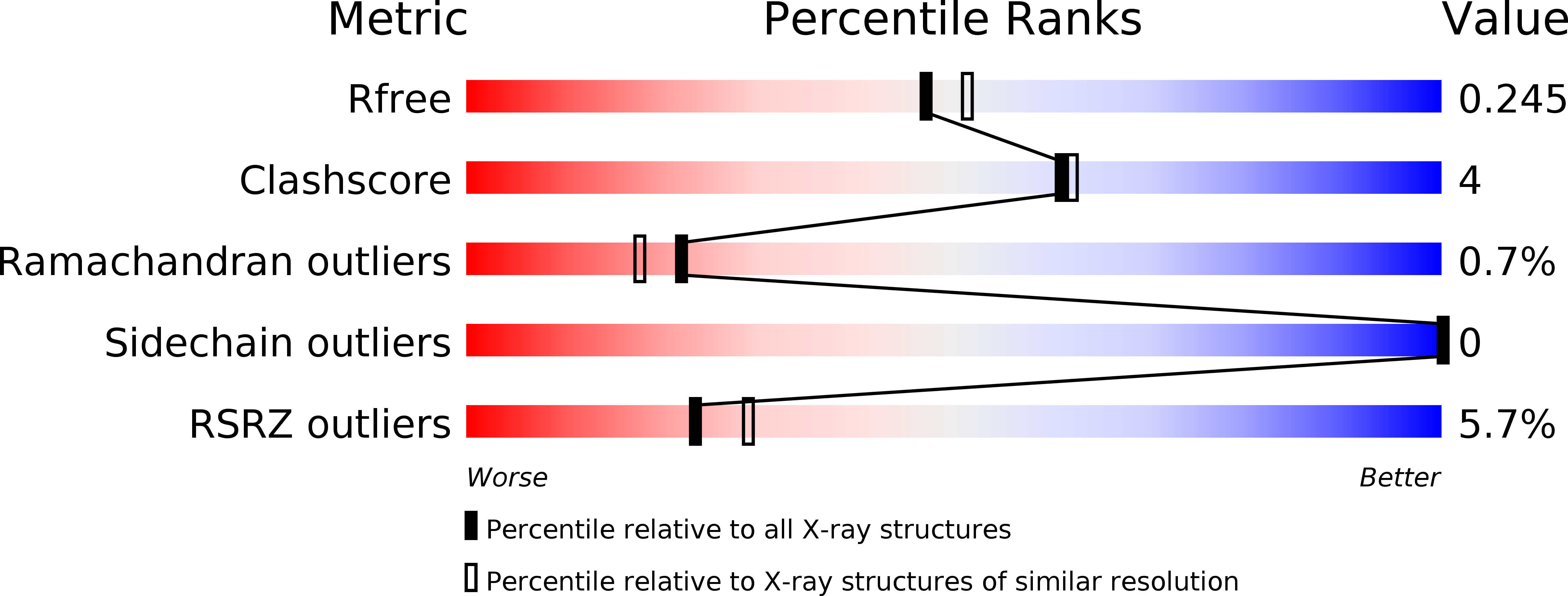
4JMF - PubMed Abstract:
ExoT belongs to the family of type 3 secretion system (T3SS) effector toxins in Pseudomonas aeruginosa, known to be one of the major virulence determinant toxins that cause chronic and acute infections in immuno-compromised individuals, burn victims and cystic fibrosis patients. Here, we report the X-ray crystal structure of the amino terminal fragment of effector toxin ExoT, in complex with full-length homodimeric chaperone SpcS at 2.1 Å resolution. The full-length dimeric chaperone SpcS has the conserved α-β-β-β-α-β-β-α fold of class I chaperones, the characteristic hydrophobic patches for binding effector proteins and a conserved polar cavity at the dimeric interface. The stable crystallized amino terminal fragment of ExoT consists of a chaperone binding domain and a membrane localization domain that wraps around the dimeric chaperone. Site-directed mutagenesis experiments and a molecular dynamics study complement each other in revealing Asn65, Phe67 and Trp88 as critical dimeric interfacial residues that can strongly influence the effector-chaperone interactions.
Organizational Affiliation:
Department of Structural Biology and Bioinformatics, Indian Institute of Chemical Biology, Kolkata, India.