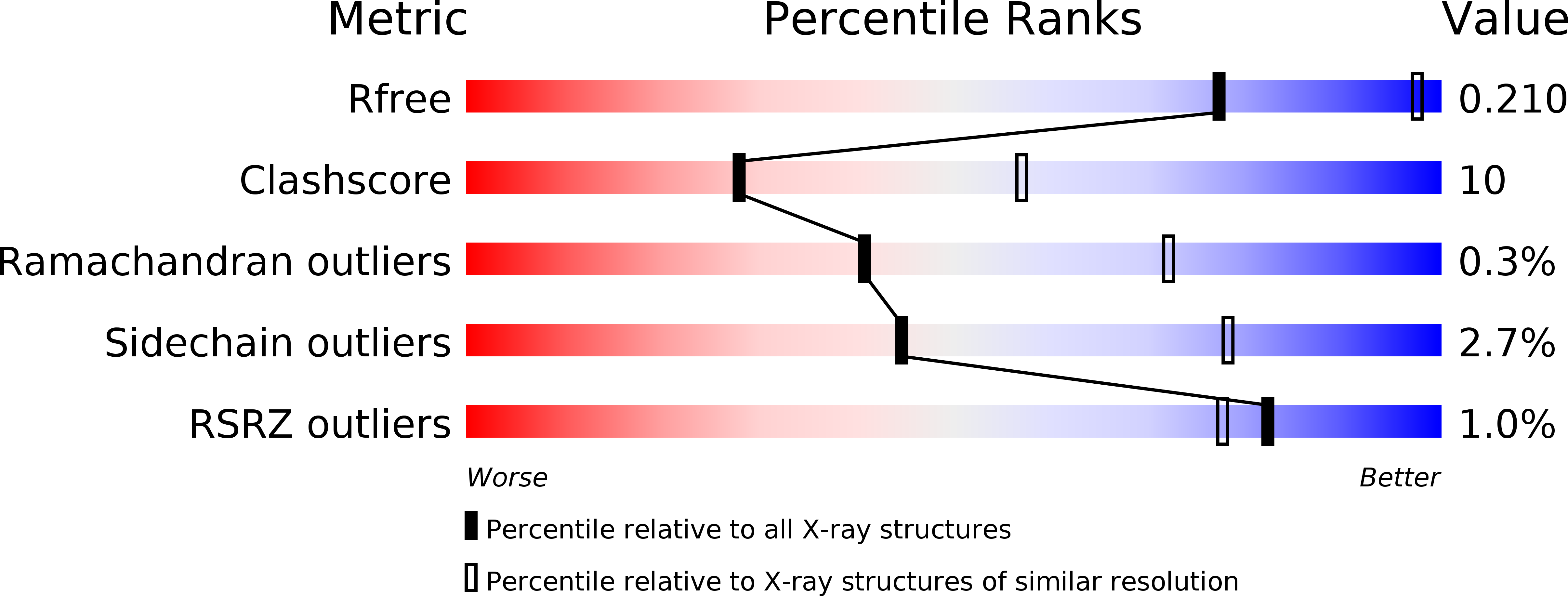
Structure of a Dimeric Crenarchaeal Cas6 Enzyme with an Atypical Active Site for Crispr RNA Processing
Reeks, J., Sokolowski, R., Graham, S., Liu, H., Naismith, J.H., White, M.F.(2013) Biochem J 452: 223
- PubMed: 23527601
- DOI: https://doi.org/10.1042/BJ20130269
- Primary Citation of Related Structures:
3ZFV - PubMed Abstract:
The competition between viruses and hosts is played out in all branches of life. Many prokaryotes have an adaptive immune system termed 'CRISPR' (clustered regularly interspaced short palindromic repeats) which is based on the capture of short pieces of viral DNA. The captured DNA is integrated into the genomic DNA of the organism flanked by direct repeats, transcribed and processed to generate crRNA (CRISPR RNA) that is loaded into a variety of effector complexes. These complexes carry out sequence-specific detection and destruction of invading mobile genetic elements. In the present paper, we report the structure and activity of a Cas6 (CRISPR-associated 6) enzyme (Sso1437) from Sulfolobus solfataricus responsible for the generation of unit-length crRNA species. The crystal structure reveals an unusual dimeric organization that is important for the enzyme's activity. In addition, the active site lacks the canonical catalytic histidine residue that has been viewed as an essential feature of the Cas6 family. Although several residues contribute towards catalysis, none is absolutely essential. Coupled with the very low catalytic rate constants of the Cas6 family and the plasticity of the active site, this suggests that the crRNA recognition and chaperone-like activities of the Cas6 family should be considered as equal to or even more important than their role as traditional enzymes.
Organizational Affiliation:
Biomedical Sciences Research Complex, University of St Andrews, North Haugh, St Andrews, Fife KY16 9ST, UK.