Structural characterization of the catalytic site of a Nilaparvata lugens delta-class glutathione transferase.
Yamamoto, K., Higashiura, A., Hossain, M.T., Yamada, N., Shiotsuki, T., Nakagawa, A.(2014) Arch Biochem Biophys 566C: 36-42
- PubMed: 25497345
- DOI: https://doi.org/10.1016/j.abb.2014.12.001
- Primary Citation of Related Structures:
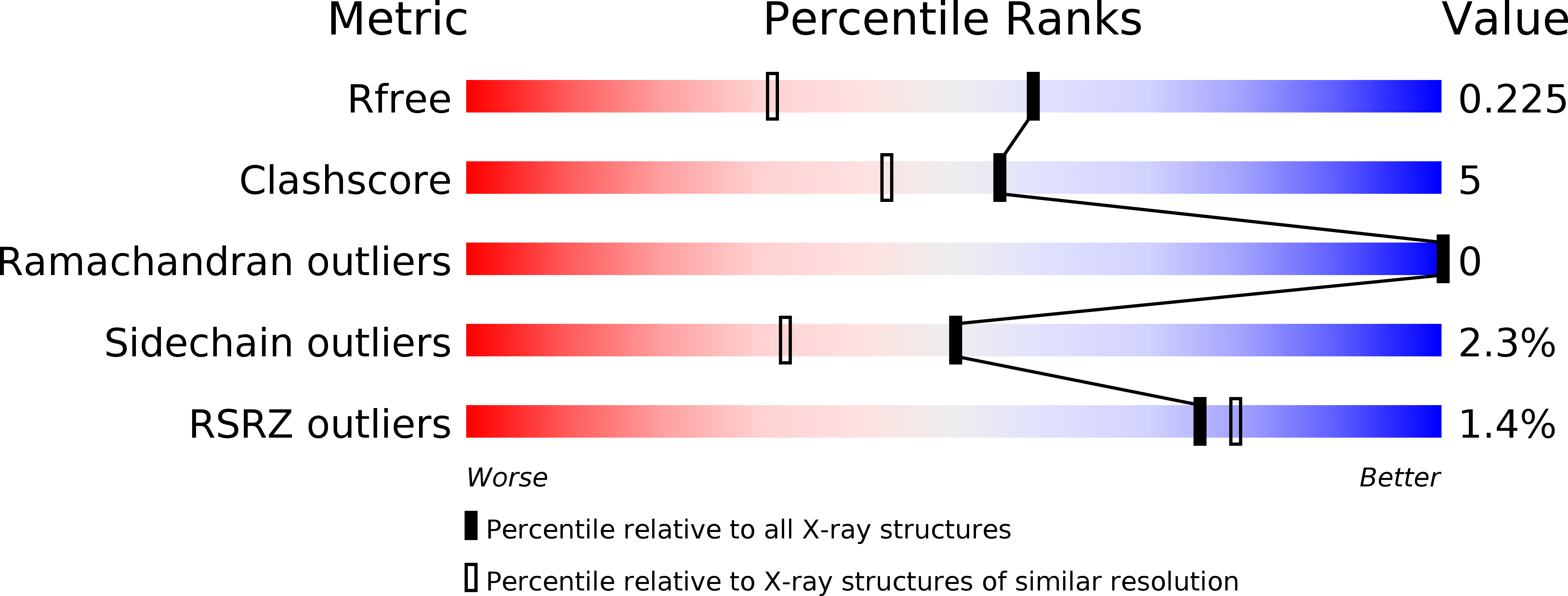
3WYW - PubMed Abstract:
Glutathione transferases (GSTs) are a major class of detoxification enzymes that play a central role in the defense against environmental toxicants and oxidative stress. Here, we studied the crystal structure of a delta-class glutathione transferase from Nilaparvata lugens, nlGSTD, to gain insights into its catalytic mechanism. The structure of nlGSTD in complex with glutathione, determined at a resolution of 1.7Å, revealed that it exists as a dimer and its secondary and tertiary structures are similar to those of other delta-class GSTs. Analysis of a complex between nlGSTD and glutathione showed that the bound glutathione was localized to the glutathione-binding site. Site-directed mutagenesis of nlGSTD mutants indicated that amino acid residues Ser11, His52, Glu66, and Phe119 contribute to catalytic activity.
Organizational Affiliation:
Faculty of Agriculture, Kyushu University Graduate School, 6-10-1 Hakozaki, Higashi-ku, Fukuoka 812-8581, Japan. Electronic address: yamamok@agr.kyushu-u.ac.jp.