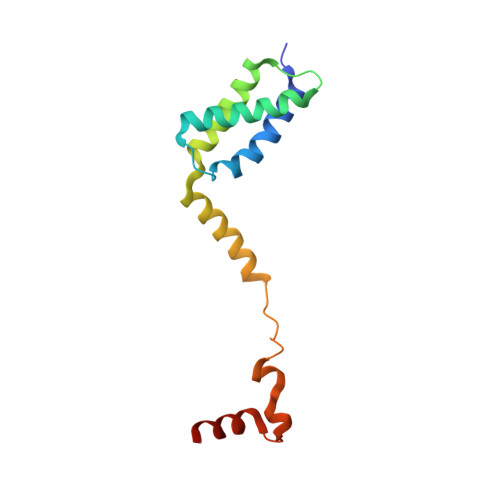
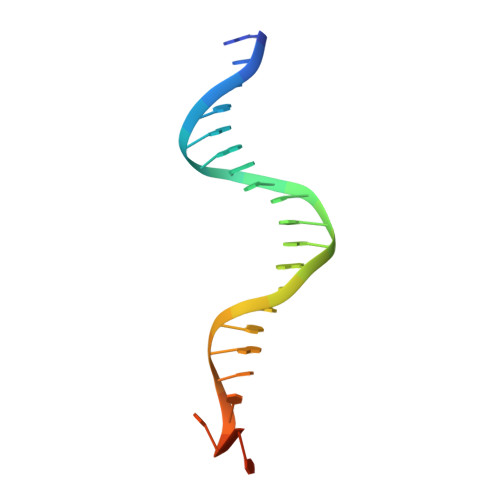
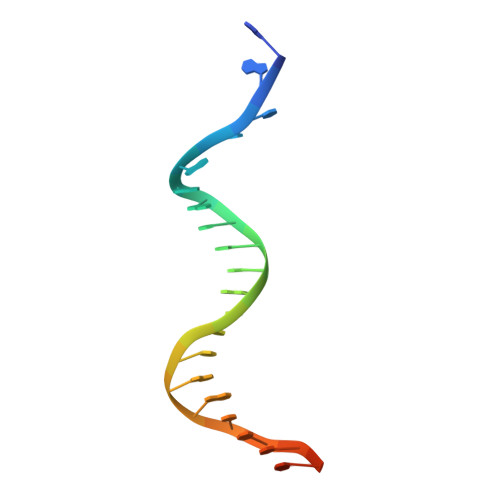
Structural Basis for Replication Origin Unwinding by An Initiator-Primase of Plasmid ColE2-P9: Duplex DNA Unwinding by A Single Protein
Itou, H., Yagura, M., Shirakihara, Y., Itoh, T.(2015) J Biol Chem 290: 3601-3611
- PubMed: 25538245
- DOI: https://doi.org/10.1074/jbc.M114.595645
- Primary Citation of Related Structures:
3VW4 - PubMed Abstract:
Duplex DNA is generally unwound by protein oligomers prior to replication. The Rep protein of plasmid ColE2-P9 (34 kDa) is an essential initiator for plasmid DNA replication. This protein binds the replication origin (Ori) in a sequence-specific manner as a monomer and unwinds DNA. Here we present the crystal structure of the DNA-binding domain of Rep (E2Rep-DBD) in complex with Ori DNA. The structure unveils the basis for Ori-specific recognition by the E2Rep-DBD and also reveals that it unwinds DNA by the concerted actions of its three contiguous structural modules. The structure also shows that the functionally unknown PriCT domain, which forms a compact module, plays a central role in DNA unwinding. The conservation of the PriCT domain in the C termini of some archaeo-eukaryotic primases indicates that it probably plays a similar role in these proteins. Thus, this is the first report providing the structural basis for the functional importance of the conserved PriCT domain and also reveals a novel mechanism for DNA unwinding by a single protein.
Organizational Affiliation:
From the Structural Biology Center, National Institute of Genetics, Yata 1111, Mishima, Shizuoka 411-8540, Japan, hitou@nig.ac.jp.