Tight Conformational Coupling between the Domains of the Enterotoxigenic Escherichia coli Fimbrial Adhesin CfaE Regulates Binding State Transition.
Liu, Y., Esser, L., Interlandi, G., Kisiela, D.I., Tchesnokova, V., Thomas, W.E., Sokurenko, E., Xia, D., Savarino, S.J.(2013) J Biol Chem 288: 9993-10001
- PubMed: 23393133
- DOI: https://doi.org/10.1074/jbc.M112.413534
- Primary Citation of Related Structures:
3VAC - PubMed Abstract:
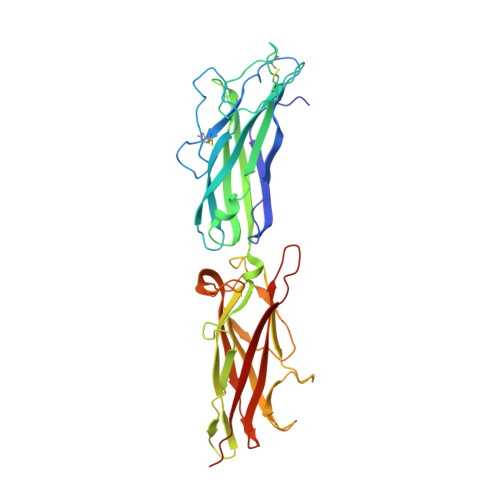
CfaE, the tip adhesin of enterotoxigenic Escherichia coli colonization factor antigen I fimbriae, initiates binding of this enteropathogen to the small intestine. It comprises stacked β-sandwich adhesin (AD) and pilin (PD) domains, with the putative receptor-binding pocket at one pole and an equatorial interdomain interface. CfaE binding to erythrocytes is enhanced by application of moderate shear stress. A G168D replacement along the AD facing the CfaE interdomain region was previously shown to decrease the dependence on shear by increasing binding at lower shear forces. To elucidate the structural basis for this functional change, we studied the properties of CfaE G168D (with a self-complemented donor strand) and solved its crystal structure at 2.6 Å resolution. Compared with native CfaE, CfaE G168D showed a downward shift in peak erythrocyte binding under shear stress and greater binding under static conditions. The thermal melting transition of CfaE G168D occurred 10 °C below that of CfaE. Compared with CfaE, the atomic structure of CfaE G168D revealed a 36% reduction in the buried surface area at the interdomain interface. Despite the location of this single modification in the AD, CfaE G168D exhibited structural derangements only in the adjoining PD compared with CfaE. In molecular dynamics simulations, the G168D mutation was associated with weakened interdomain interactions under tensile force. Taken together, these findings indicate that the AD and PD of CfaE are conformationally tightly coupled and support the hypothesis that opening of the interface plays a critical modulatory role in the allosteric activation of CfaE.
Organizational Affiliation:
Enteric Diseases Department, Infectious Diseases Directorate, Naval Medical Research Center, Silver Spring, Maryland 20910.