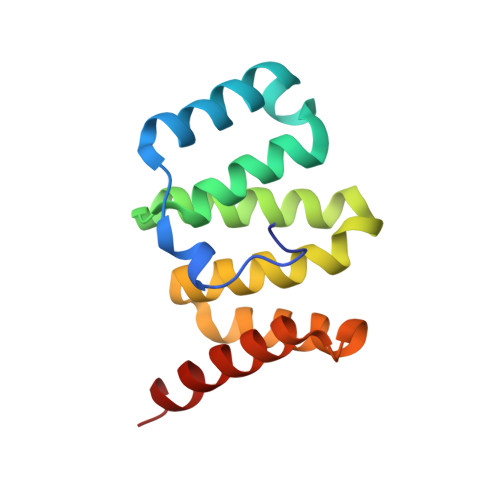
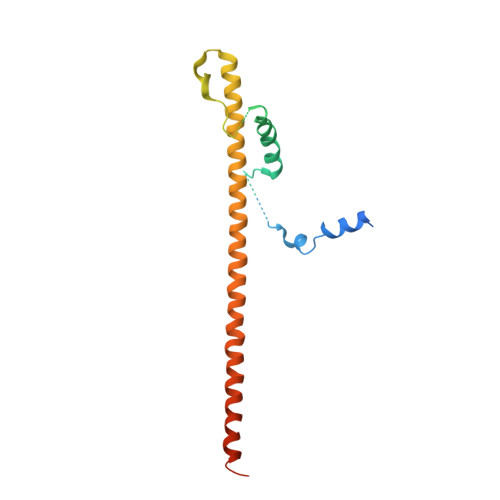
Crystal structure of mitochondrial fission complex reveals scaffolding function for mitochondrial division 1 (mdv1) coiled coil.
Zhang, Y., Chan, N.C., Ngo, H.B., Gristick, H., Chan, D.C.(2012) J Biol Chem 287: 9855-9861
- PubMed: 22303011
- DOI: https://doi.org/10.1074/jbc.M111.329359
- Primary Citation of Related Structures:
3UUX - PubMed Abstract:
The mitochondrial fission machinery is best understood in the yeast Saccharomyces cerevisiae, where Fis1, Mdv1, and Dnm1 are essential components. Fis1 is a mitochondrial outer membrane protein that recruits the dynamin-related GTPase Dnm1 during the fission process. This recruitment occurs via Mdv1, which binds both Fis1 and Dnm1 and therefore functions as a molecular adaptor linking the two molecules. Mdv1 has a modular structure, consisting of an N-terminal extension that binds Fis1, a central coiled coil for dimerization, and a C-terminal WD40 repeat region that binds Dnm1. We have solved the crystal structure of a dimeric Mdv1-Fis1 complex that contains both the N-terminal extension and coiled-coil regions of Mdv1. Consistent with previous studies, Mdv1 binds Fis1 through a U-shaped helix-loop-helix motif, and dimerization of the Mdv1-Fis1 complex is mediated by the antiparallel coiled coil of Mdv1. However, the complex is surprisingly compact and rigid due to two additional contacts mediated by the surface of the Mdv1 coiled coil. The coiled coil packs against both Fis1 and the second helix of the Mdv1 helix-loop-helix motif. Mutational analyses showed that these contacts are important for mitochondrial fission activity. These results indicate that, in addition to dimerization, the unusually long Mdv1 coiled coil serves a scaffolding function to stabilize the Mdv1-Fis1 complex.
Organizational Affiliation:
Division of Biology, California Institute of Technology, Pasadena, California 91125.