Improvement of Thermal Stability via Outer-Loop Ion Pair Interaction of Mutated T1 Lipase from Geobacillus zalihae Strain T1
Ruslan, R., Rahman, R.N.Z.R.A., Leow, T.C., Ali, M.S.M., Basri, M., Salleh, A.B.(2012) Int J Mol Sci 13: 943-960
- PubMed: 22312296
- DOI: https://doi.org/10.3390/ijms13010943
- Primary Citation of Related Structures:
3UMJ - PubMed Abstract:
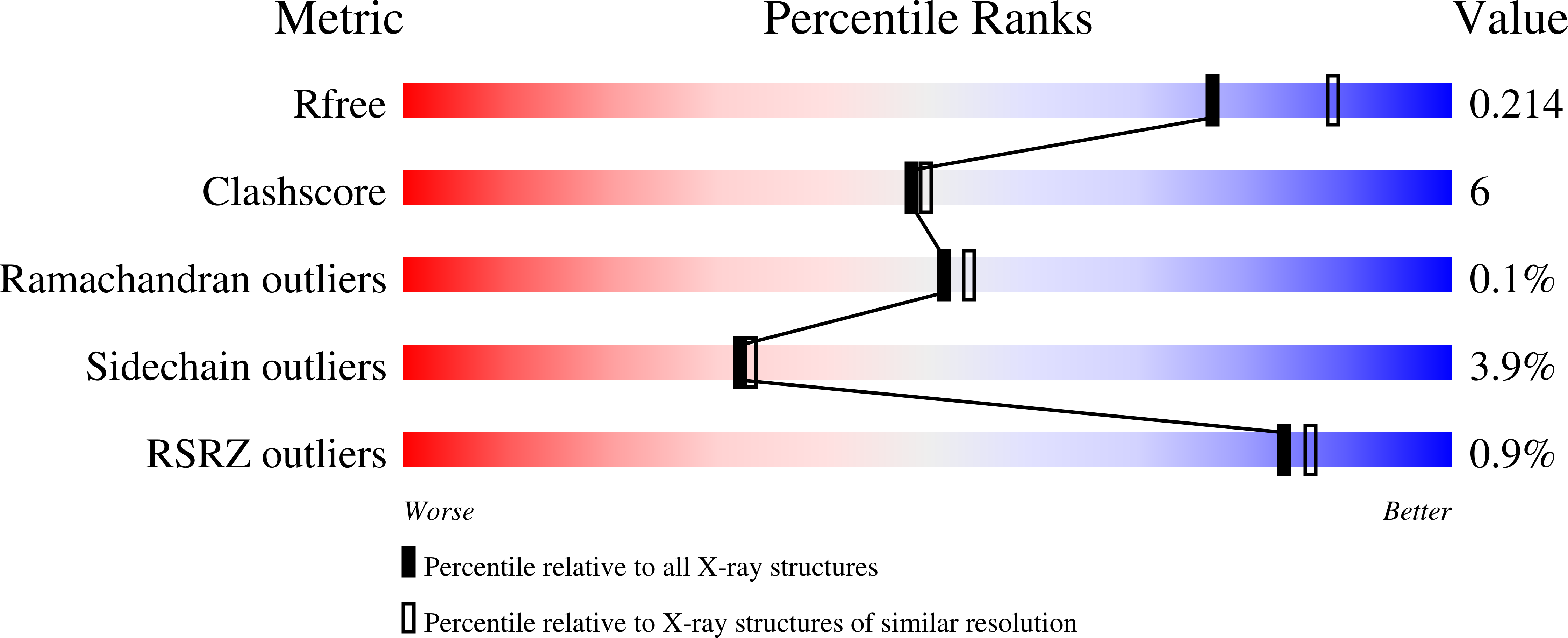
Mutant D311E and K344R were constructed using site-directed mutagenesis to introduce an additional ion pair at the inter-loop and the intra-loop, respectively, to determine the effect of ion pairs on the stability of T1 lipase isolated from Geobacillus zalihae. A series of purification steps was applied, and the pure lipases of T1, D311E and K344R were obtained. The wild-type and mutant lipases were analyzed using circular dichroism. The T(m) for T1 lipase, D311E lipase and K344R lipase were approximately 68.52 °C, 70.59 °C and 68.54 °C, respectively. Mutation at D311 increases the stability of T1 lipase and exhibited higher T(m) as compared to the wild-type and K344R. Based on the above, D311E lipase was chosen for further study. D311E lipase was successfully crystallized using the sitting drop vapor diffusion method. The crystal was diffracted at 2.1 Å using an in-house X-ray beam and belonged to the monoclinic space group C2 with the unit cell parameters a = 117.32 Å, b = 81.16 Å and c = 100.14 Å. Structural analysis showed the existence of an additional ion pair around E311 in the structure of D311E. The additional ion pair in D311E may regulate the stability of this mutant lipase at high temperatures as predicted in silico and spectroscopically.
Organizational Affiliation:
Enzyme and Microbial Technology Laboratory, Faculty of Biotechnology and Biomolecular Sciences, Universiti Putra Malaysia, 43400 UPM Serdang, Selangor, Malaysia.