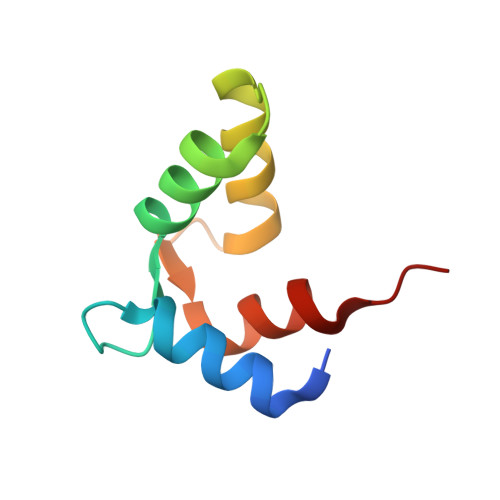
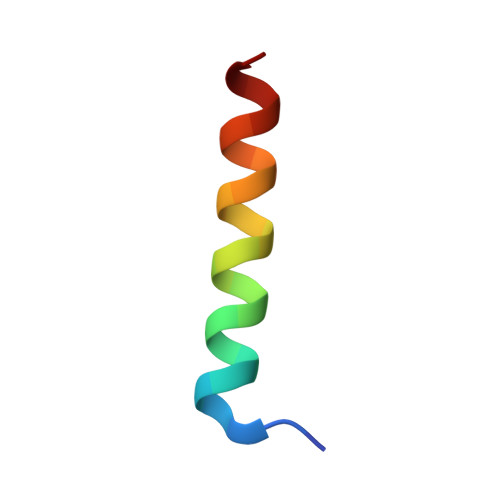
Structure of the Ca2+-saturated C-terminal domain of scallop troponin C in complex with a troponin I fragment
Kato, Y.S., Yumoto, F., Tanaka, H., Miyakawa, T., Miyauchi, Y., Takeshita, D., Sawano, Y., Ojima, T., Ohtsuki, I., Tanokura, M.(2012) Biol Chem 394: 55-68
- PubMed: 23096565
- DOI: https://doi.org/10.1515/hsz-2012-0152
- Primary Citation of Related Structures:
3TZ1 - PubMed Abstract:
Troponin C (TnC) is the Ca(2+)-sensing subunit of troponin that triggers the contraction of striated muscles. In scallops, the striated muscles consume little ATP energy in sustaining strong contractile forces. The N-terminal domain of TnC works as the Ca(2+) sensor in vertebrates, whereas scallop TnC uses the C-terminal domain as the Ca(2+) sensor, suggesting that there are differences in the mechanism of the Ca(2+)-dependent regulation of muscles between invertebrates and vertebrates. Here, we report the crystal structure of Akazara scallop (Chlamys nipponensis akazara) adductor muscle TnC C-terminal domain (asTnCC) complexed with a short troponin I fragment (asTnIS) and Ca(2+). The electron density of a Ca(2+) ion is observed in only one of the two EF-hands. The EF-hands of asTnCC can only be in the fully open conformation with the assistance of asTnIS. The number of hydrogen bonds between asTnCC and asTnIS is markedly lower than the number in the vertebrate counterparts. The Ca(2+) modulation on the binding between asTnCC and asTnIS is weaker, but structural change of the complex depending on Ca(2+) concentration was observed. Together, these findings provide a detailed description of the distinct molecular mechanism of contractile regulation in the scallop adductor muscle from that of vertebrates.
Organizational Affiliation:
Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo 113-8657, Japan.