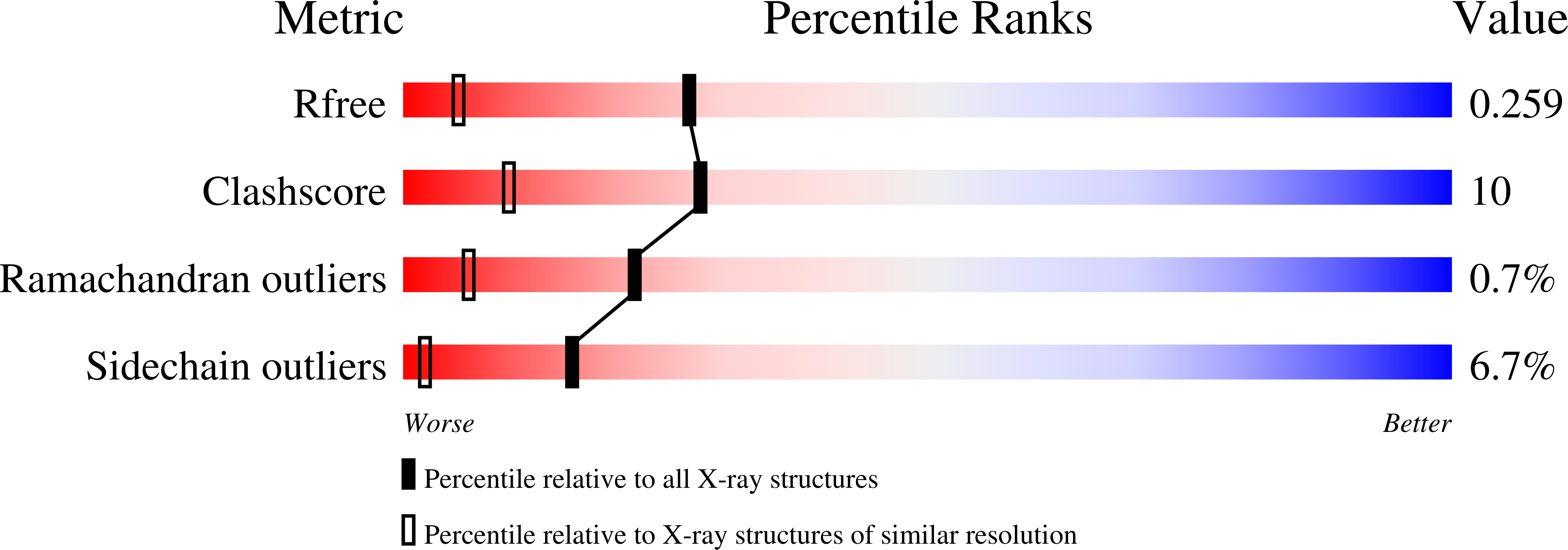
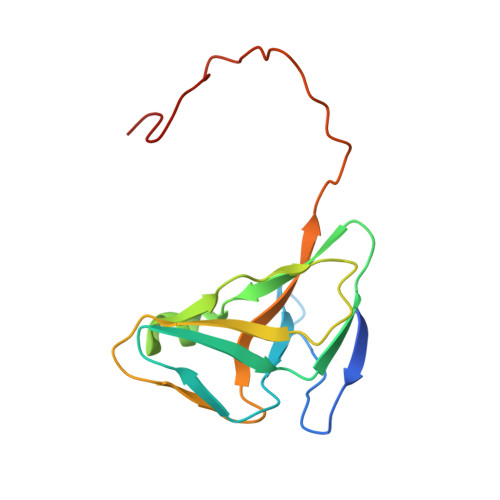
Crystallization of Chlorella deoxyuridine triphosphatase.
Badalucco, L., Poudel, I., Yamanishi, M., Natarajan, C., Moriyama, H.(2011) Acta Crystallogr Sect F Struct Biol Cryst Commun 67: 1599-1602
- PubMed: 22139176
- DOI: https://doi.org/10.1107/S1744309111038097
- Primary Citation of Related Structures:
3SO2 - PubMed Abstract:
Deoxyuridine triphosphatase (dUTPase) is a ubiquitous enzyme that has been widely studied owing to its function and evolutionary significance. The gene coding for the dUTPase from the Chlorella alga was codon-optimized and synthesized. The synthetic gene was expressed in Escherichia coli and recombinant core Chlorella dUTPase (chdUTPase) was purified. Crystallization of chdUTPase was performed by the repetitive hanging-drop vapor-diffusion method at 298 K with ammonium sulfate as the precipitant. In the presence of 2'-deoxyuridine-5'-[(α,β)-imido]triphosphate and magnesium, the enzyme produced die-shaped hexagonal R3 crystals with unit-cell parameters a = b = 66.9, c = 93.6 Å, γ = 120°. X-ray diffraction data for chdUTPase were collected to 1.6 Å resolution. The crystallization of chdUTPase with manganese resulted in very fragile clusters of needles.
Organizational Affiliation:
School of Biological Sciences, University of Nebraska-Lincoln, Lincoln, NE 68588-0118, USA.