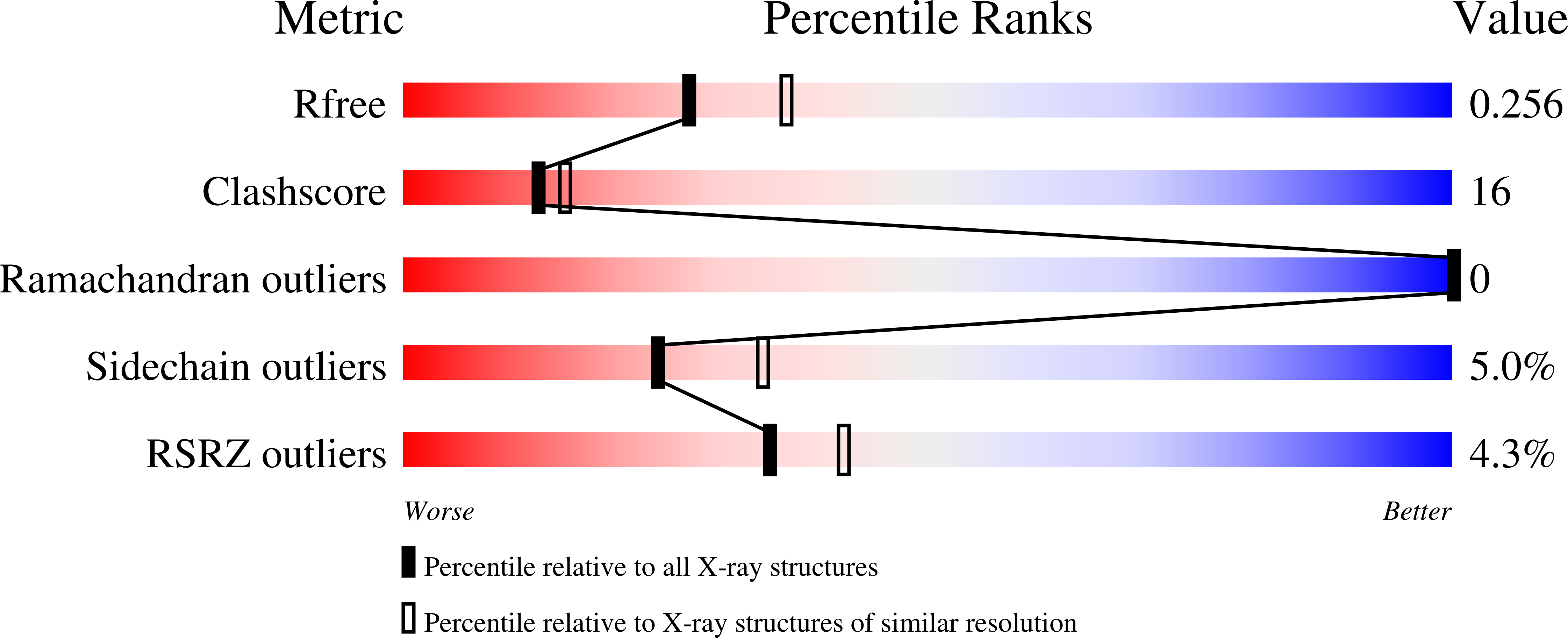
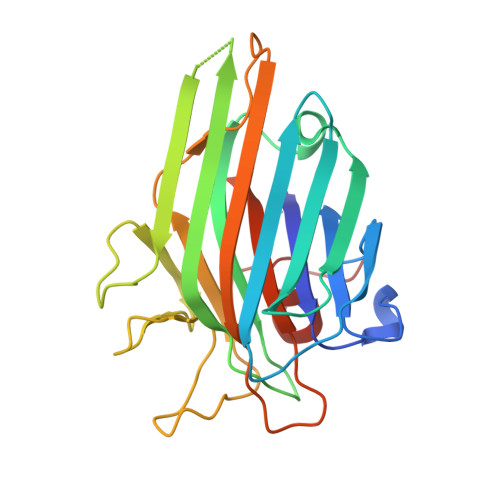
Crystal structure of a pro-inflammatory lectin from the seeds of Dioclea wilsonii Standl.
Rangel, T.B., Rocha, B.A., Bezerra, G.A., Assreuy, A.M., Pires, A.F., Nascimento, A.S., Bezerra, M.J., Nascimento, K.S., Nagano, C.S., Sampaio, A.H., Gruber, K., Delatorre, P., Fernandes, P.M., Cavada, B.S.(2012) Biochimie 94: 525-532
- PubMed: 21924319
- DOI: https://doi.org/10.1016/j.biochi.2011.09.001
- Primary Citation of Related Structures:
3SH3 - PubMed Abstract:
The crystal structure and pro-inflammatory property of a lectin from the seeds of Dioclea wilsonii (DwL) were analyzed to gain a better understanding of structure/function relationships of Diocleinae lectins. Following crystallization and structural determination by standard molecular replacement techniques, DwL was found to be a tetramer based on PISA analysis, and composed by two metal-binding sites per monomer and loops which are involved in molecular oligomerization. DwL presents 96% and 99% identity with two other previously described lectins of Dioclea rostrata (DRL) and Dioclea grandiflora (DGL). DwL differs structurally from DVL and DRL with regard to the conformation of the carbohydrate recognition domain and related biological activities. The structural analysis of DwL in comparison to other Diocleinae lectins can be related to the differences in the dose-dependent pro-inflammatory effect elicited in Wistar rats, probably via specific interactions with mast cells complex carbohydrate, resulting in significant paw edema. DwL appears to be involved in positive modulation of mast cell degranulation via recognition of surface carbohydrates. Since this recognition is dependent on site volume and CRD configuration, edematogenesis mediated by resident cells varies in potency and efficacy among different Diocleinae lectins.
Organizational Affiliation:
Núcleo de Biotecnologia, Universidade Federal do Espírito Santo, Vitória, Brazil.