Structural basis of regiospecificity of a mononuclear iron enzyme in antibiotic fosfomycin biosynthesis.
Yun, D., Dey, M., Higgins, L.J., Yan, F., Liu, H.W., Drennan, C.L.(2011) J Am Chem Soc 133: 11262-11269
- PubMed: 21682308
- DOI: https://doi.org/10.1021/ja2025728
- Primary Citation of Related Structures:
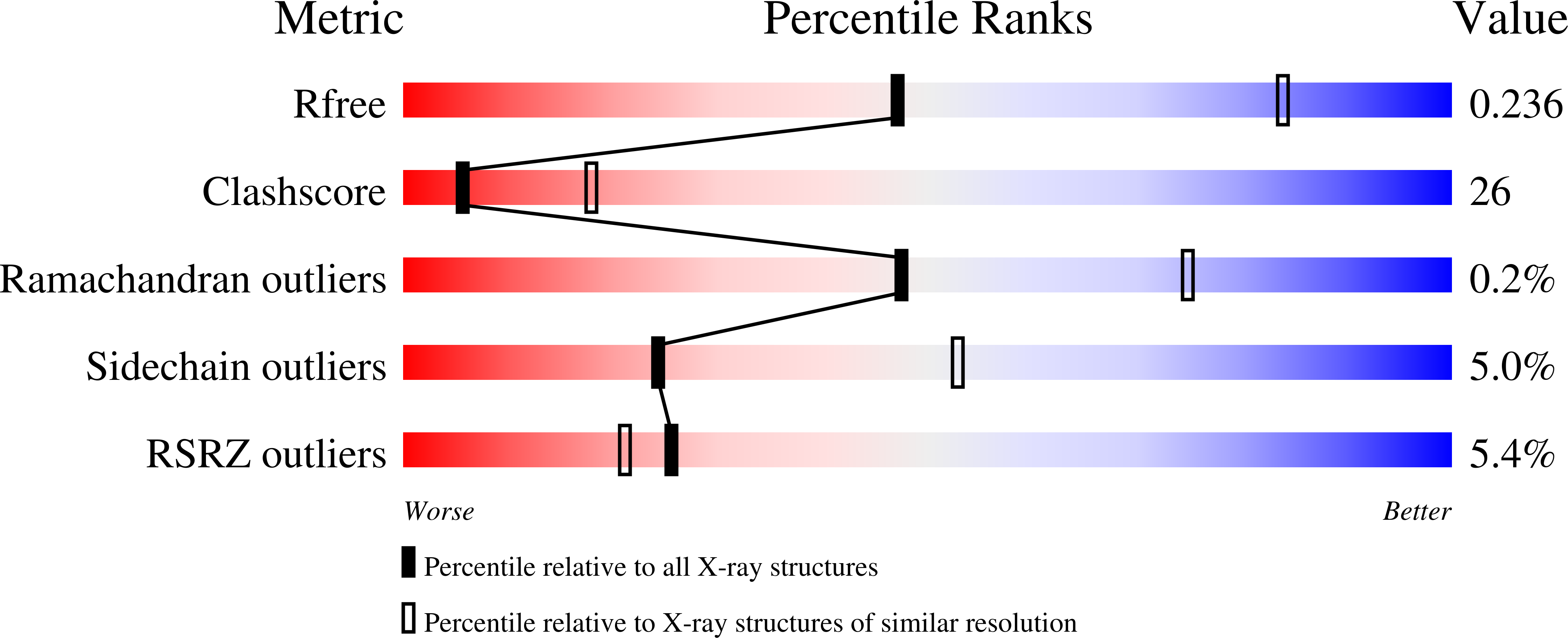
3SCF, 3SCG, 3SCH - PubMed Abstract:
Hydroxypropylphosphonic acid epoxidase (HppE) is an unusual mononuclear iron enzyme that uses dioxygen to catalyze the oxidative epoxidation of (S)-2-hydroxypropylphosphonic acid (S-HPP) in the biosynthesis of the antibiotic fosfomycin. Additionally, the enzyme converts the R-enantiomer of the substrate (R-HPP) to 2-oxo-propylphosphonic acid. To probe the mechanism of HppE regiospecificity, we determined three X-ray structures: R-HPP with inert cobalt-containing enzyme (Co(II)-HppE) at 2.1 Å resolution; R-HPP with active iron-containing enzyme (Fe(II)-HppE) at 3.0 Å resolution; and S-HPP-Fe(II)-HppE in complex with dioxygen mimic NO at 2.9 Å resolution. These structures, along with previously determined structures of S-HPP-HppE, identify the dioxygen binding site on iron and elegantly illustrate how HppE is able to recognize both substrate enantiomers to catalyze two completely distinct reactions.
Organizational Affiliation:
Department of Chemistry, Massachusetts Institute of Technology, Cambridge, Massachusetts 02139, USA.