Synthesis and characterization of the arylomycin lipoglycopeptide antibiotics and the crystallographic analysis of their complex with signal peptidase.
Liu, J., Luo, C., Smith, P.A., Chin, J.K., Page, M.G., Paetzel, M., Romesberg, F.E.(2011) J Am Chem Soc 133: 17869-17877
- PubMed: 21999324
- DOI: https://doi.org/10.1021/ja207318n
- Primary Citation of Related Structures:
3S04 - PubMed Abstract:
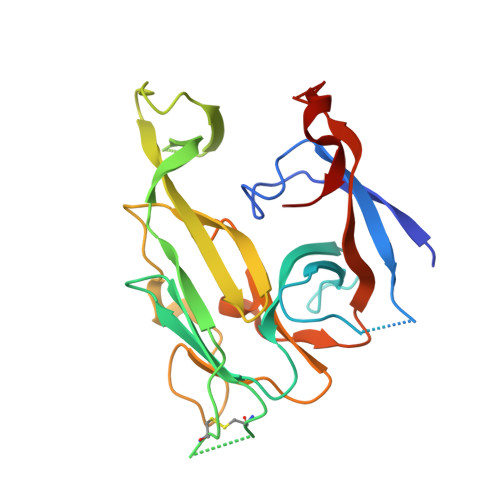
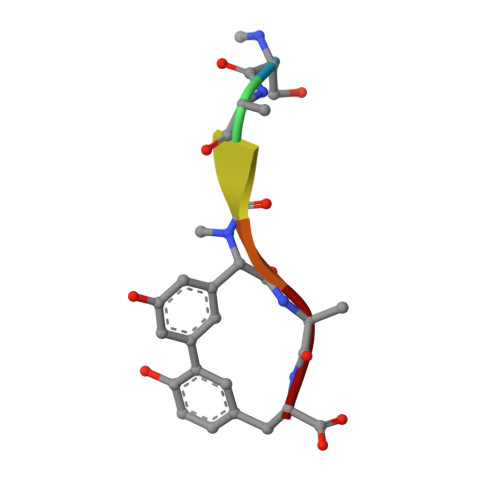
Glycosylation of natural products, including antibiotics, often plays an important role in determining their physical properties and their biological activity, and thus their potential as drug candidates. The arylomycin class of antibiotics inhibits bacterial type I signal peptidase and is comprised of three related series of natural products with a lipopeptide tail attached to a core macrocycle. Previously, we reported the total synthesis of several A series derivatives, which have unmodified core macrocycles, as well as B series derivatives, which have a nitrated macrocycle. We now report the synthesis and biological evaluation of lipoglycopeptide arylomycin variants whose macrocycles are glycosylated with a deoxy-α-mannose substituent, and also in some cases hydroxylated. The synthesis of the derivatives bearing each possible deoxy-α-mannose enantiomer allowed us to assign the absolute stereochemistry of the sugar in the natural product and also to show that while glycosylation does not alter antibacterial activity, it does appear to improve solubility. Crystallographic structural studies of a lipoglycopeptide arylomycin bound to its signal peptidase target reveal the molecular interactions that underlie inhibition and also that the mannose is directed away from the binding site into solvent which suggests that other modifications may be made at the same position to further increase solubility and thus reduce protein binding and possibly optimize the pharmacokinetics of the scaffold.
Organizational Affiliation:
Department of Chemistry, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, California 92037, United States.