Kinetic, Crystallographic, and Mechanistic Characterization of TomN: Elucidation of a Function for a 4-Oxalocrotonate Tautomerase Homologue in the Tomaymycin Biosynthetic Pathway.
Burks, E.A., Yan, W., Johnson, W.H., Li, W., Schroeder, G.K., Min, C., Gerratana, B., Zhang, Y., Whitman, C.P.(2011) Biochemistry 50: 7600-7611
- PubMed: 21809870
- DOI: https://doi.org/10.1021/bi200947w
- Primary Citation of Related Structures:
3RY0 - PubMed Abstract:
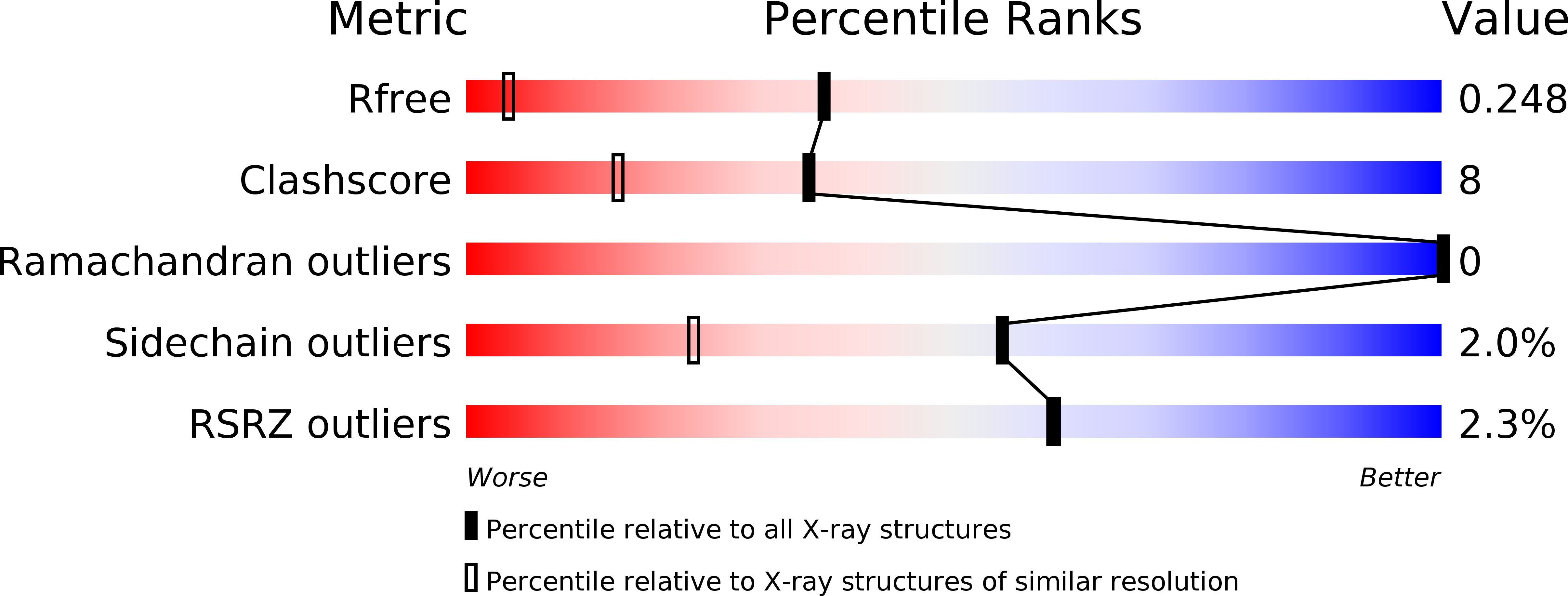
The biosynthesis of the C ring of the antitumor antibiotic agent, tomaymycin, is proposed to proceed through five enzyme-catalyzed steps from l-tyrosine. The genes encoding these enzymes have recently been cloned and their functions tentatively assigned, but there is limited biochemical evidence supporting the assignments of the last three steps. One enzyme, TomN, shows 58% pairwise sequence similarity with 4-oxalocrotonate tautomerase (4-OT), an enzyme found in a catabolic pathway for aromatic hydrocarbons. The TomN sequence includes three amino acids (Pro-1, Arg-11, and Arg-39) that have been identified as critical catalytic residues in 4-OT. However, the proposed substrate for TomN is very different from that processed by 4-OT. To establish the function and mechanism of TomN and its relationship with 4-OT, we conducted kinetic, mutagenic, and structural studies. The kinetic parameters for TomN, and four alanine mutants, P1A, R11A, R39A, and R61A, were determined using 2-hydroxymuconate, the substrate for 4-OT. The TomN-catalyzed reaction using this substrate compares favorably to that of 4-OT. In addition, the kinetic parameters for the P1A, R11A, and R39A mutants of TomN parallel the trends observed for the corresponding 4-OT mutants, implicating an analogous mechanism. A high-resolution crystal structure (1.4 Å) of TomN shows that the overall structure and the active site region are highly similar to those of 4-OT with a root-mean-square deviation of 0.81 Å. Moreover, key active site residues are positionally conserved. The combined results suggest that the tentative assignment for TomN and the proposed sequence of events in the biosynthetic pathway leading to the formation of the C ring of tomaymycin might not be correct. An alternative pathway that awaits biochemical confirmation is proposed.
Organizational Affiliation:
Division of Medicinal Chemistry, College of Pharmacy, University of Texas, Austin, Texas 78712, United States.