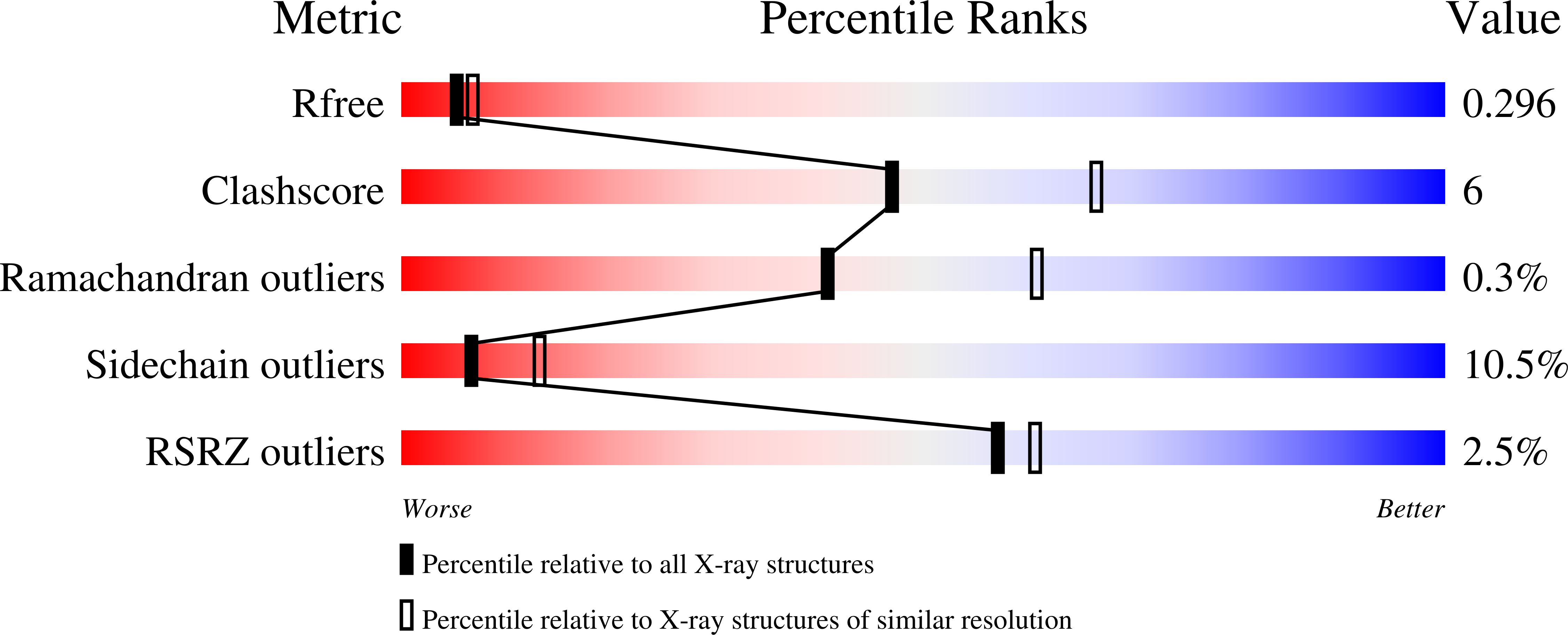
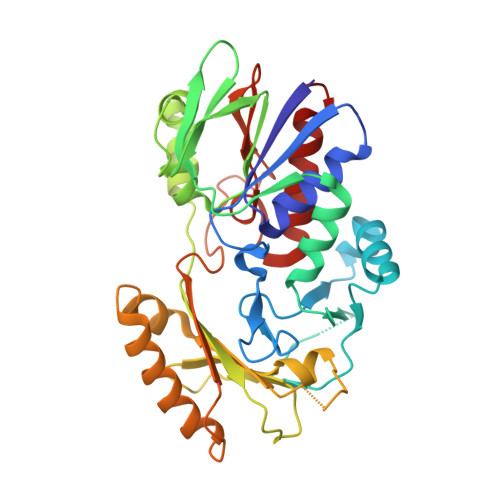
FAD-binding site and NADP reactivity in human renalase: a new enzyme involved in blood pressure regulation
Milani, M., Ciriello, F., Baroni, S., Pandini, V., Canevari, G., Bolognesi, M., Aliverti, A.(2011) J Mol Biol 411: 463-473
- PubMed: 21699903
- DOI: https://doi.org/10.1016/j.jmb.2011.06.010
- Primary Citation of Related Structures:
3QJ4 - PubMed Abstract:
Renalase is a recently discovered flavoprotein that regulates blood pressure, regulates sodium and phosphate excretion, and displays cardioprotectant action through a mechanism that is barely understood to date. It has been proposed to act as a catecholamine-degrading enzyme, via either O(2)-dependent or NADH-dependent mechanisms. Here we report the renalase crystal structure at 2.5 Å resolution together with new data on its interaction with nicotinamide dinucleotides. Renalase adopts the p-hydroxybenzoate hydroxylase fold topology, comprising a Rossmann-fold-based flavin adenine dinucleotide (FAD)-binding domain and a putative substrate-binding domain, the latter of which contains a five-stranded anti-parallel β-sheet. A large cavity (228 Å(3)), facing the flavin ring, presumably represents the active site. Compared to monoamine oxidase or polyamine oxidase, the renalase active site is fully solvent exposed and lacks an 'aromatic cage' for binding the substrate amino group. Renalase has an extremely low diaphorase activity, displaying lower k(cat) but higher k(cat)/K(m) for NADH compared to NADPH. Moreover, its FAD prosthetic group becomes slowly reduced when it is incubated with NADPH under anaerobiosis, and binds NAD(+) or NADP(+) with K(d) values of ca 2 mM. The absence of a recognizable NADP-binding site in the protein structure and its poor affinity for, and poor reactivity towards, NADH and NADPH suggest that these are not physiological ligands of renalase. Although our study does not answer the question on the catalytic activity of renalase, it provides a firm framework for testing hypotheses on the molecular mechanism of its action.
Organizational Affiliation:
Dipartimento di Scienze Biomolecolari e Biotecnologie, Università degli Studi di Milano, via Celoria 26, 20133 Milan, Italy.