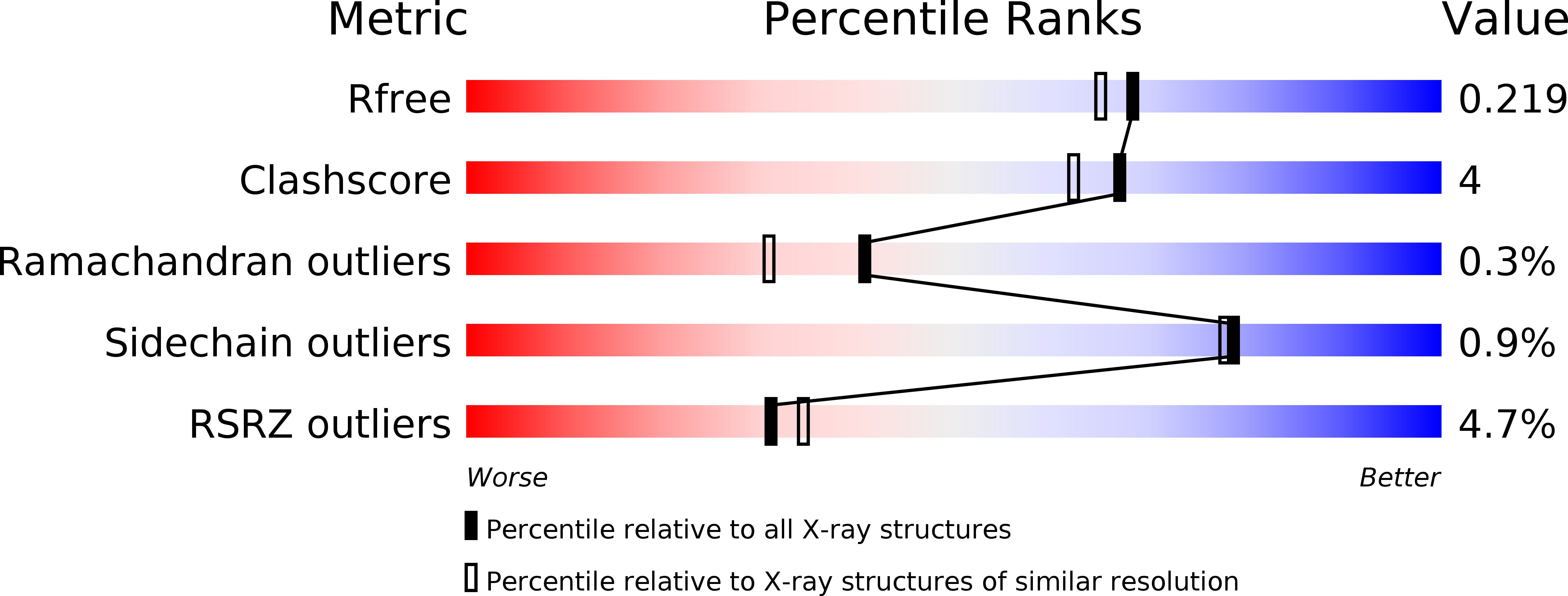
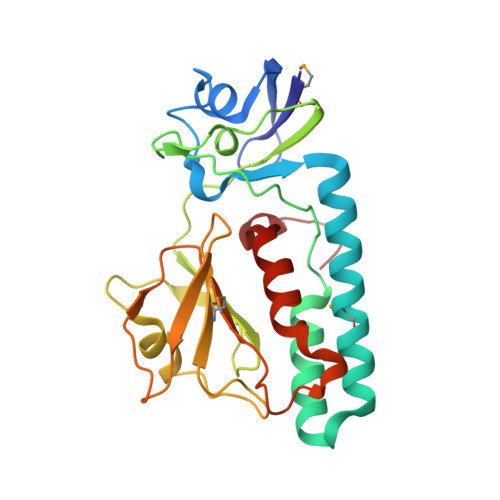
The Crystal Structure of Escherichia coli Group 4 Capsule Protein GfcC Reveals a Domain Organization Resembling That of Wza.
Sathiyamoorthy, K., Mills, E., Franzmann, T.M., Rosenshine, I., Saper, M.A.(2011) Biochemistry 50: 5465-5476
- PubMed: 21449614
- DOI: https://doi.org/10.1021/bi101869h
- Primary Citation of Related Structures:
3P42 - PubMed Abstract:
We report the 1.9 Å resolution crystal structure of enteropathogenic Escherichia coli GfcC, a periplasmic protein encoded by the gfc operon, which is essential for assembly of group 4 polysaccharide capsule (O-antigen capsule). Presumed gene orthologs of gfcC are present in capsule-encoding regions of at least 29 genera of Gram-negative bacteria. GfcC, a member of the DUF1017 family, is comprised of tandem β-grasp (ubiquitin-like) domains (D2 and D3) and a carboxyl-terminal amphipathic helix, a domain arrangement reminiscent of that of Wza that forms an exit pore for group 1 capsule export. Unlike the membrane-spanning C-terminal helix from Wza, the GfcC C-terminal helix packs against D3. Previously unobserved in a β-grasp domain structure is a 48-residue helical hairpin insert in D2 that binds to D3, constraining its position and sequestering the carboxyl-terminal amphipathic helix. A centrally located and invariant Arg115 not only is essential for proper localization but also forms one of two mostly conserved pockets. Finally, we draw analogies between a GfcC protein fused to an outer membrane β-barrel pore in some species and fusion proteins necessary for secreting biofilm-forming exopolysaccharides.
Organizational Affiliation:
Program in Biophysics, University of Michigan, Ann Arbor, Michigan 48109-1055, USA.