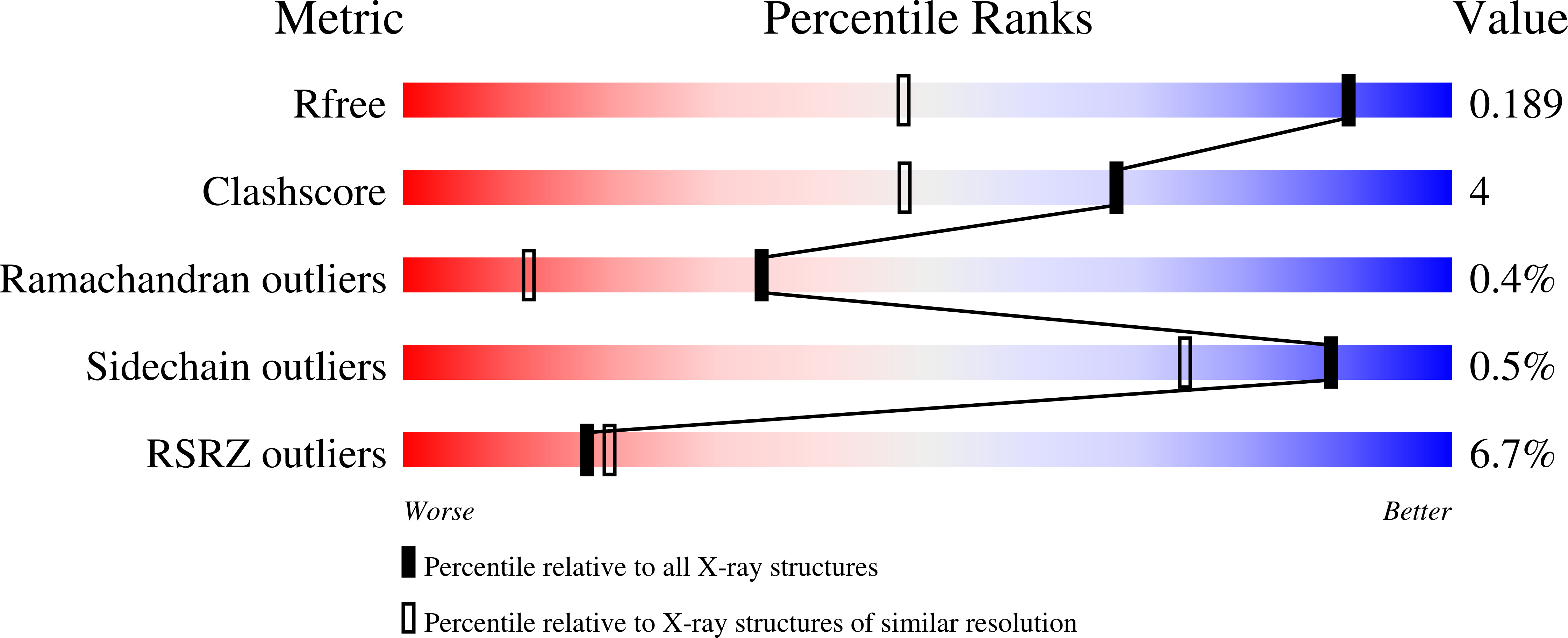
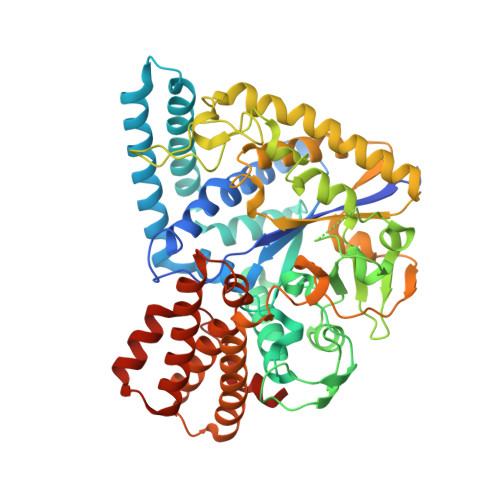
Thermus thermophilus GLYCOSYL HYDROLASE FAMILY 57 branching enzyme: crystal structure, mechanism of action and products formed
Palomo-Reixach, M., Pijning, T., Booiman, T., Dobruchowska, J., van der Vlist, J., Kralj, S., Planas, A., Loos, K., Kamerling, J.P., Dijkstra, B.W., van der Maarel, M.J.E.C., Dijkhuizen, L., Leemhuis, H.To be published.