In vitro and in vivo studies of the trypanocidal properties of WRR-483 against Trypanosoma cruzi.
Chen, Y.T., Brinen, L.S., Kerr, I.D., Hansell, E., Doyle, P.S., McKerrow, J.H., Roush, W.R.(2010) PLoS Negl Trop Dis 4
- PubMed: 20856868
- DOI: https://doi.org/10.1371/journal.pntd.0000825
- Primary Citation of Related Structures:
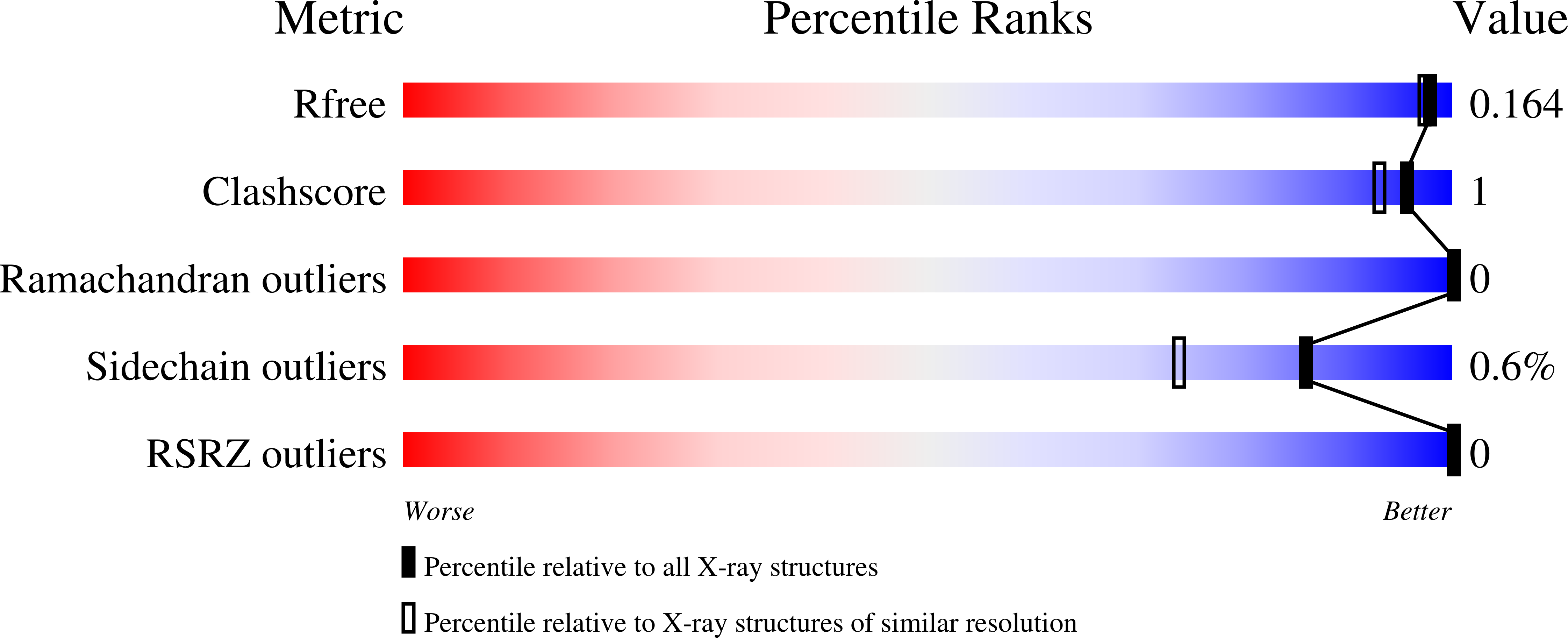
3LXS - PubMed Abstract:
Cruzain, the major cysteine protease of Trypanosoma cruzi, is an essential enzyme for the parasite life cycle and has been validated as a viable target to treat Chagas' disease. As a proof-of-concept, K11777, a potent inhibitor of cruzain, was found to effectively eliminate T. cruzi infection and is currently a clinical candidate for treatment of Chagas' disease.
Organizational Affiliation:
Department of Chemistry, The Scripps Research Institute, Scripps Florida, Jupiter, Florida, United States of America.