Crystal structure of the cystic fibrosis transmembrane conductance regulator inhibitory factor Cif reveals novel active-site features of an epoxide hydrolase virulence factor.
Bahl, C.D., Morisseau, C., Bomberger, J.M., Stanton, B.A., Hammock, B.D., O'Toole, G.A., Madden, D.R.(2010) J Bacteriol 192: 1785-1795
- PubMed: 20118260
- DOI: https://doi.org/10.1128/JB.01348-09
- Primary Citation of Related Structures:
3KD2, 3KDA - PubMed Abstract:
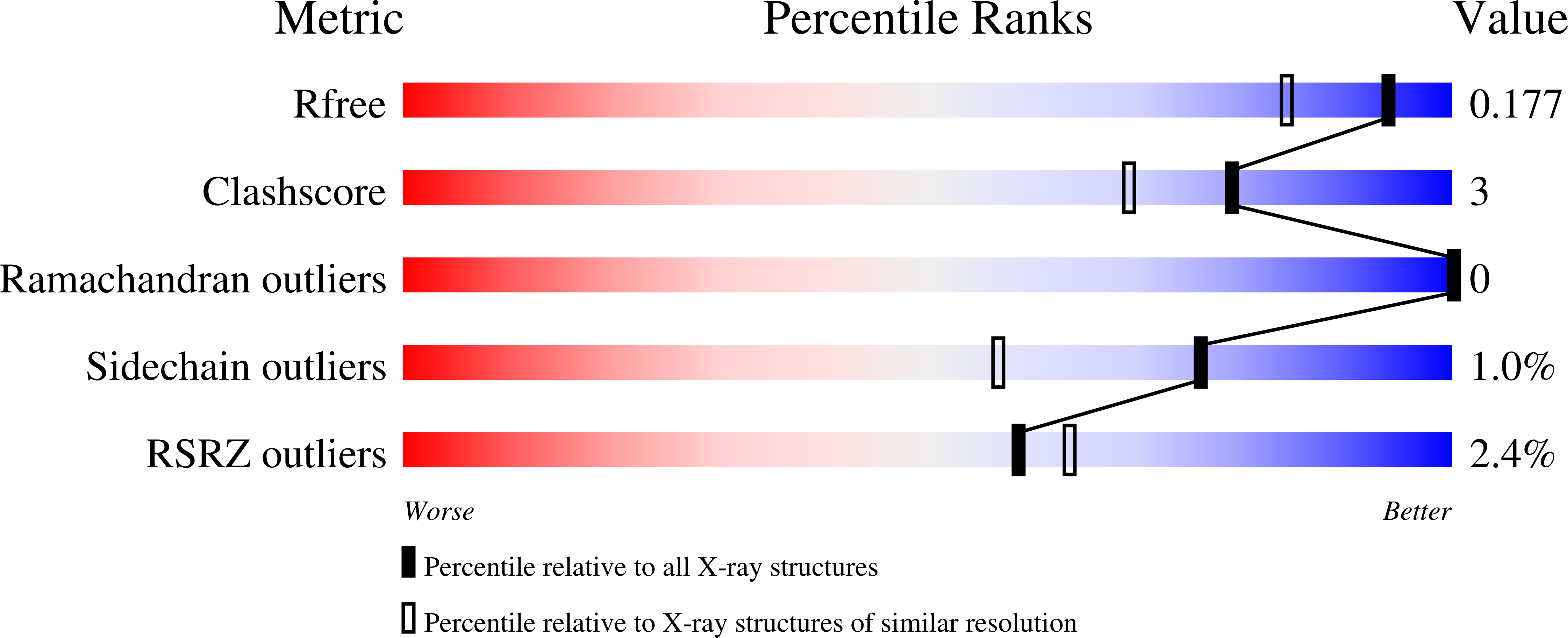
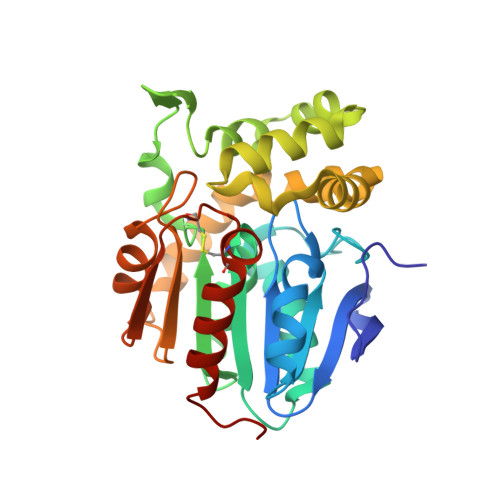
Cystic fibrosis transmembrane conductance regulator (CFTR) inhibitory factor (Cif) is a virulence factor secreted by Pseudomonas aeruginosa that reduces the quantity of CFTR in the apical membrane of human airway epithelial cells. Initial sequence analysis suggested that Cif is an epoxide hydrolase (EH), but its sequence violates two strictly conserved EH motifs and also is compatible with other alpha/beta hydrolase family members with diverse substrate specificities. To investigate the mechanistic basis of Cif activity, we have determined its structure at 1.8-A resolution by X-ray crystallography. The catalytic triad consists of residues Asp129, His297, and Glu153, which are conserved across the family of EHs. At other positions, sequence deviations from canonical EH active-site motifs are stereochemically conservative. Furthermore, detailed enzymatic analysis confirms that Cif catalyzes the hydrolysis of epoxide compounds, with specific activity against both epibromohydrin and cis-stilbene oxide, but with a relatively narrow range of substrate selectivity. Although closely related to two other classes of alpha/beta hydrolase in both sequence and structure, Cif does not exhibit activity as either a haloacetate dehalogenase or a haloalkane dehalogenase. A reassessment of the structural and functional consequences of the H269A mutation suggests that Cif's effect on host-cell CFTR expression requires the hydrolysis of an extended endogenous epoxide substrate.
Organizational Affiliation:
Department of Biochemistry, Dartmouth Medical School, Hanover, NH 03755, USA.