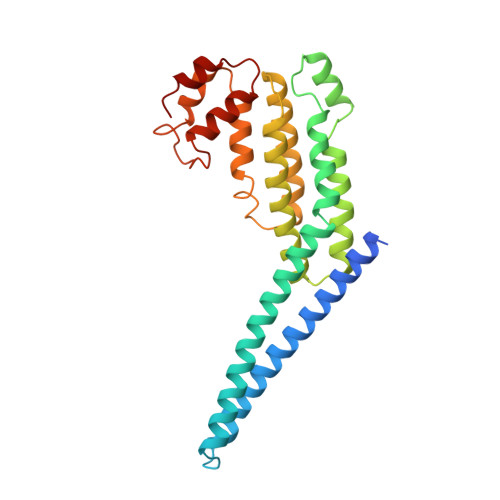
Structural basis for a human glycosylation disorder caused by mutation of the COG4 gene.
Richardson, B.C., Smith, R.D., Ungar, D., Nakamura, A., Jeffrey, P.D., Lupashin, V.V., Hughson, F.M.(2009) Proc Natl Acad Sci U S A 106: 13329-13334
- PubMed: 19651599
- DOI: https://doi.org/10.1073/pnas.0901966106
- Primary Citation of Related Structures:
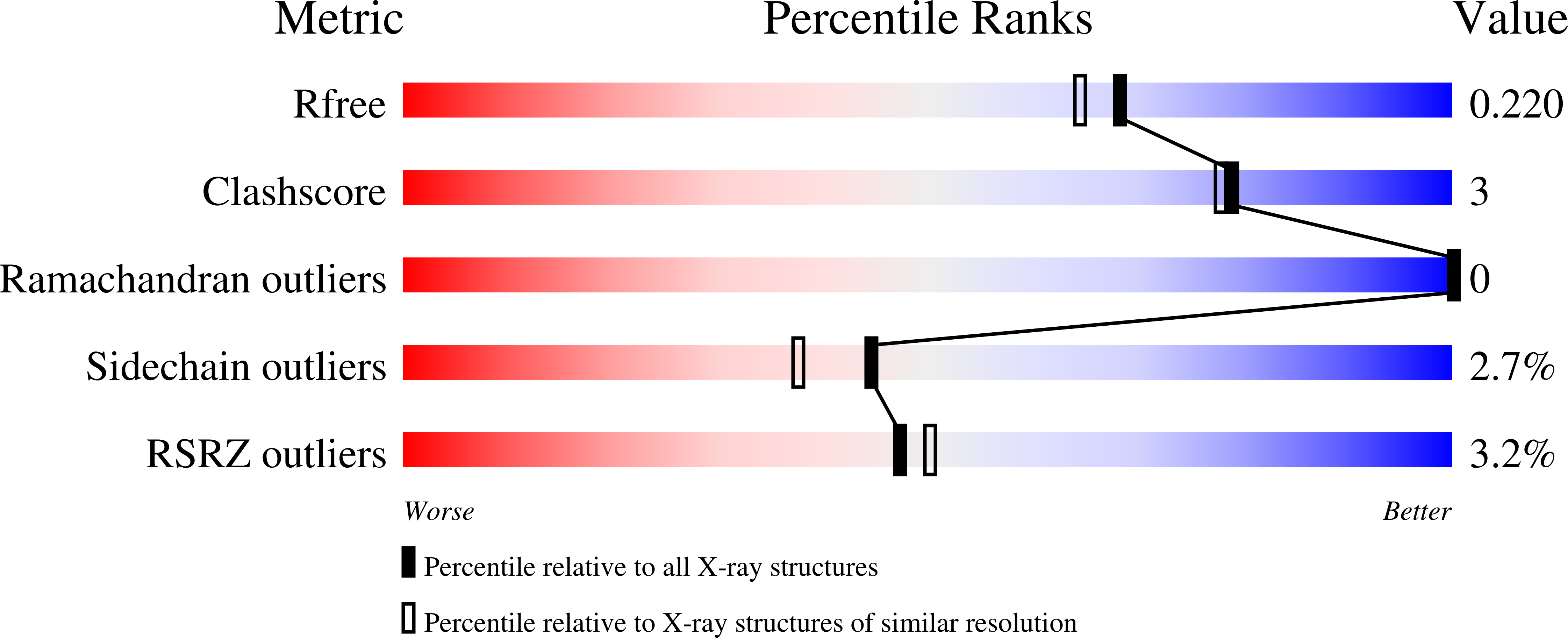
3HR0 - PubMed Abstract:
The proper glycosylation of proteins trafficking through the Golgi apparatus depends upon the conserved oligomeric Golgi (COG) complex. Defects in COG can cause fatal congenital disorders of glycosylation (CDGs) in humans. The recent discovery of a form of CDG, caused in part by a COG4 missense mutation changing Arg 729 to Trp, prompted us to determine the 1.9 A crystal structure of a Cog4 C-terminal fragment. Arg 729 is found to occupy a key position at the center of a salt bridge network, thereby stabilizing Cog4's small C-terminal domain. Studies in HeLa cells reveal that this C-terminal domain, while not needed for the incorporation of Cog4 into COG complexes, is essential for the proper glycosylation of cell surface proteins. We also find that Cog4 bears a strong structural resemblance to exocyst and Dsl1p complex subunits. These complexes and others have been proposed to function by mediating the initial tethering between transport vesicles and their membrane targets; the emerging structural similarities provide strong evidence of a common evolutionary origin and may reflect shared mechanisms of action.
Organizational Affiliation:
Department of Molecular Biology, Princeton University, Princeton, NJ 08544, USA.