KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C.
Graef, T., Moesta, A.K., Norman, P.J., Abi-Rached, L., Vago, L., Older Aguilar, A.M., Gleimer, M., Hammond, J.A., Guethlein, L.A., Bushnell, D.A., Robinson, P.J., Parham, P.(2009) J Exp Med 206: 2557-2572
- PubMed: 19858347
- DOI: https://doi.org/10.1084/jem.20091010
- Primary Citation of Related Structures:
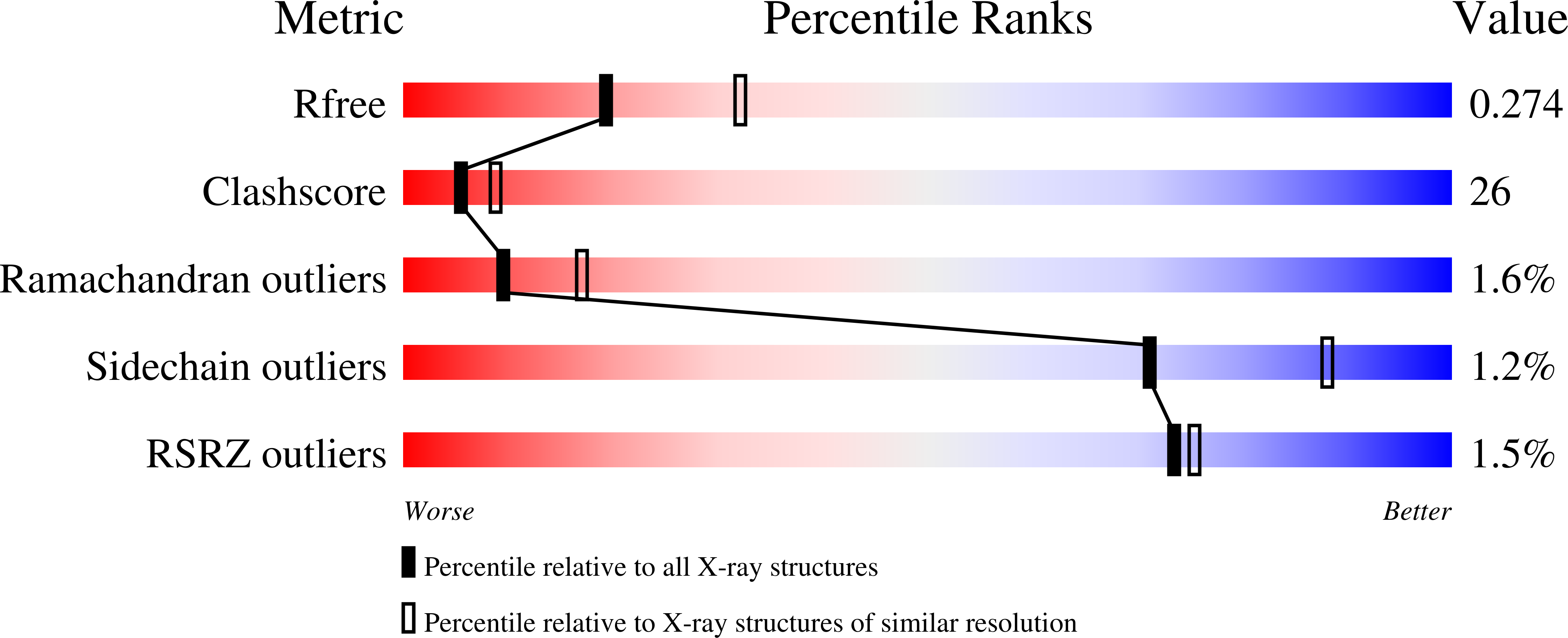
3H8N - PubMed Abstract:
Human killer cell immunoglobulin-like receptors (KIRs) are distinguished by expansion of activating KIR2DS, whose ligands and functions remain poorly understood. The oldest, most prevalent KIR2DS is KIR2DS4, which is represented by a variable balance between "full-length" and "deleted" forms. We find that full-length 2DS4 is a human histocompatibility leukocyte antigen (HLA) class I receptor that binds specifically to subsets of C1+ and C2+ HLA-C and to HLA-A*11, whereas deleted 2DS4 is nonfunctional. Activation of 2DS4+ NKL cells was achieved with A*1102 as ligand, which differs from A*1101 by unique substitution of lysine 19 for glutamate, but not with A*1101 or HLA-C. Distinguishing KIR2DS4 from other KIR2DS is the proline-valine motif at positions 71-72, which is shared with KIR3DL2 and was introduced by gene conversion before separation of the human and chimpanzee lineages. Site-directed swap mutagenesis shows that these two residues are largely responsible for the unique HLA class I specificity of KIR2DS4. Determination of the crystallographic structure of KIR2DS4 shows two major differences from KIR2DL: displacement of contact loop L2 and altered bonding potential because of the substitutions at positions 71 and 72. Correlation between the worldwide distributions of functional KIR2DS4 and HLA-A*11 points to the physiological importance of their mutual interaction.
Organizational Affiliation:
Department of Structural Biology, Stanford University School of Medicine, Stanford, CA 94305, USA.