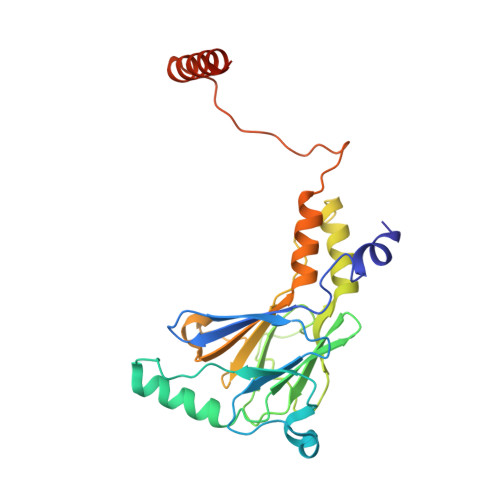
Insights into interferon regulatory factor activation from the crystal structure of dimeric IRF5.
Chen, W., Lam, S.S., Srinath, H., Jiang, Z., Correia, J.J., Schiffer, C.A., Fitzgerald, K.A., Lin, K., Royer, W.E.(2008) Nat Struct Mol Biol 15: 1213-1220
- PubMed: 18836453
- DOI: https://doi.org/10.1038/nsmb.1496
- Primary Citation of Related Structures:
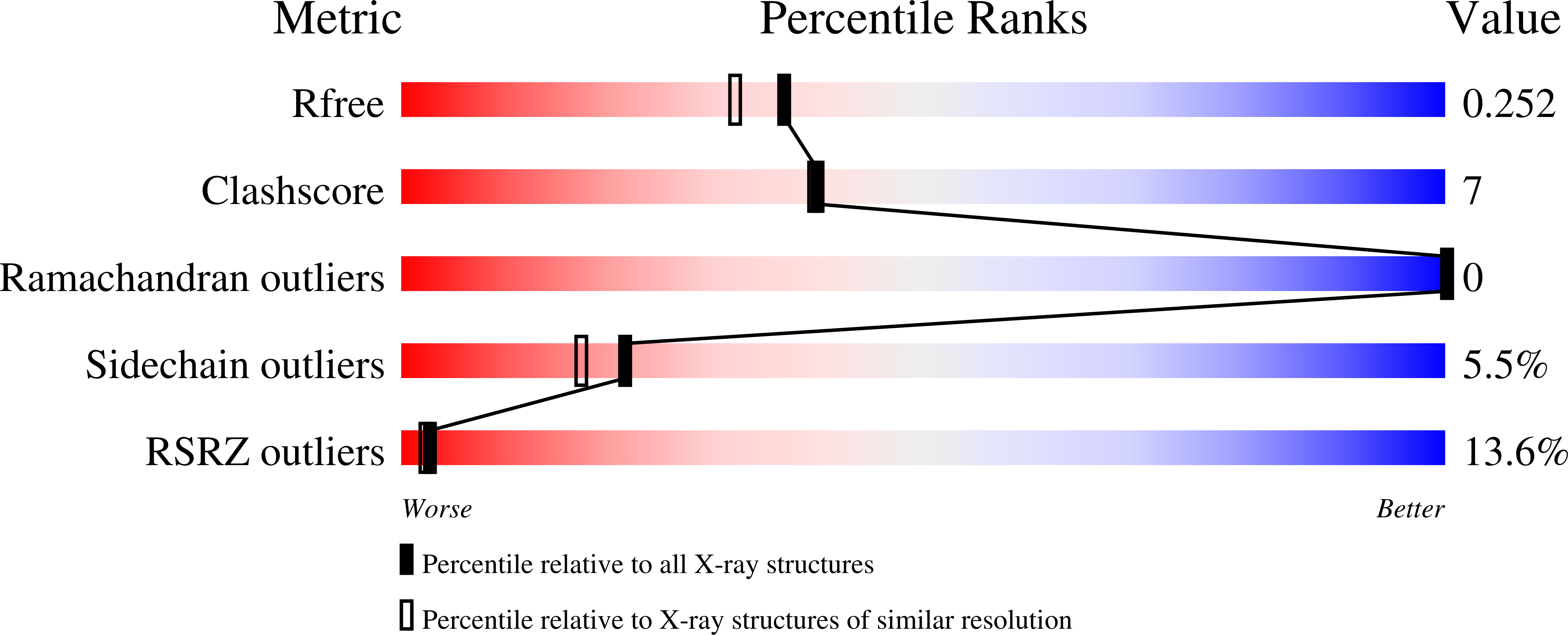
3DSH - PubMed Abstract:
Interferon regulatory factors (IRFs) are essential in the innate immune response and other physiological processes. Activation of these proteins in the cytoplasm is triggered by phosphorylation of serine and threonine residues in a C-terminal autoinhibitory region, which stimulates dimerization, transport into the nucleus, assembly with the coactivator CBP/p300 and initiation of transcription. The crystal structure of the transactivation domain of pseudophosphorylated human IRF5 strikingly reveals a dimer in which the bulk of intersubunit interactions involve a highly extended C-terminal region. The corresponding region has previously been shown to block CBP/p300 binding to unphosphorylated IRF3. Mutation of key interface residues supports the observed dimer as the physiologically activated state of IRF5 and IRF3. Thus, phosphorylation is likely to activate IRF5 and other family members by triggering conformational rearrangements that switch the C-terminal segment from an autoinihibitory to a dimerization role.
Organizational Affiliation:
Department of Biochemistry and Molecular Pharmacology, 364 Plantation Street, Worcester, Massachusetts 01605, USA.