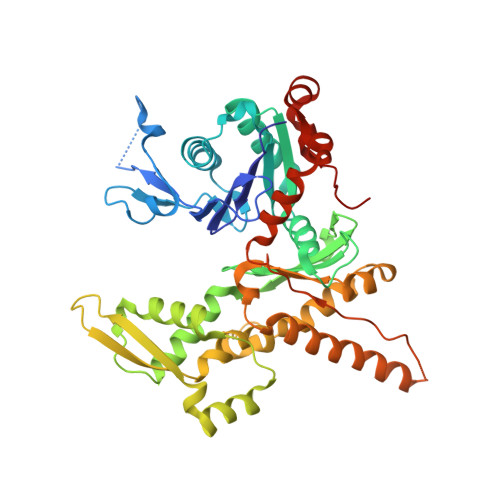
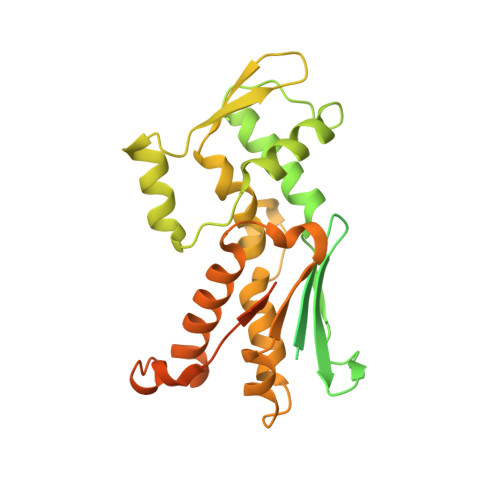
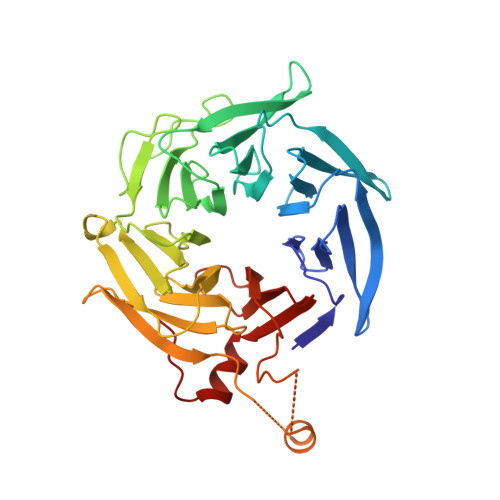
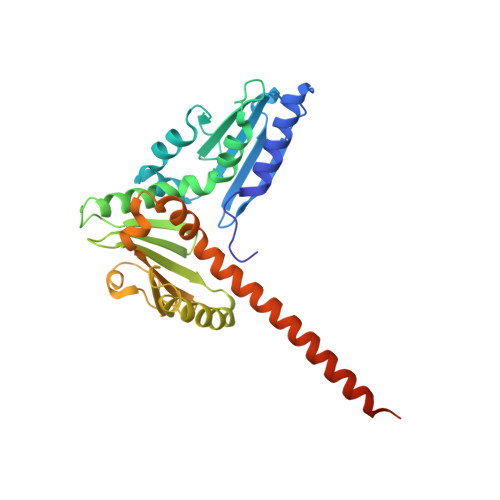
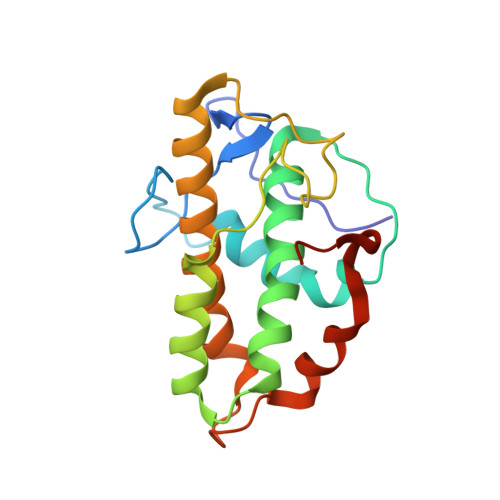
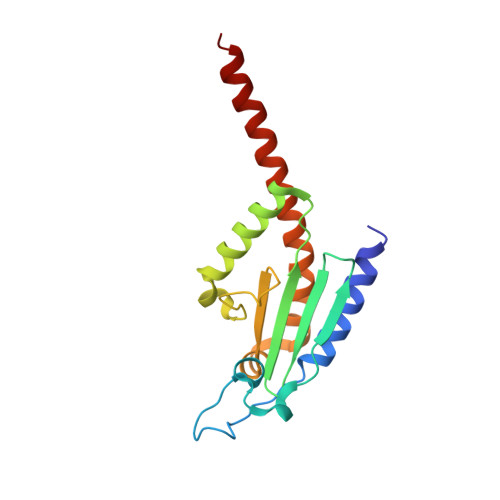
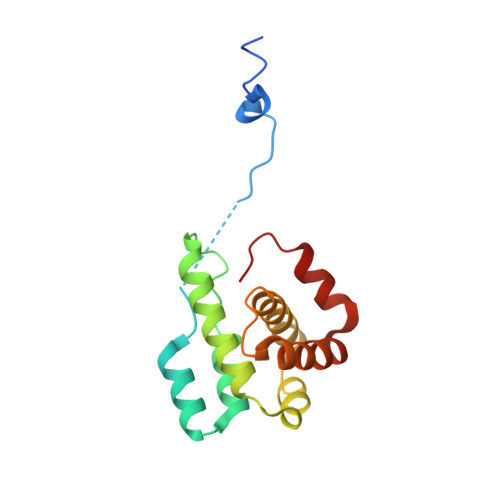
Structural characterization and computer-aided optimization of a small-molecule inhibitor of the Arp2/3 complex, a key regulator of the actin cytoskeleton.
Baggett, A.W., Cournia, Z., Han, M.S., Patargias, G., Glass, A.C., Liu, S.Y., Nolen, B.J.(2012) ChemMedChem 7: 1286-1294
- PubMed: 22623398
- DOI: https://doi.org/10.1002/cmdc.201200104
- Primary Citation of Related Structures:
3UKR - PubMed Abstract:
CK-666 (1) is a recently discovered small-molecule inhibitor of the actin-related protein 2/3 (Arp2/3) complex, a key actin cytoskeleton regulator with roles in bacterial pathogenesis and cancer cell motility. Although 1 is commercially available, the crystal structure of Arp2/3 complex with 1 bound has not been reported, making its mechanism of action uncertain. Furthermore, its relatively low potency increases its potential for off-target effects in vivo, complicating interpretation of its influence in cell biological studies and precluding its clinical use. Herein we report the crystal structure of 1 bound to Arp2/3 complex, which reveals that 1 binds between the Arp2 and Arp3 subunits to stabilize the inactive conformation of the complex. Based on the crystal structure, we used computational docking and free-energy perturbation calculations of monosubstituted derivatives of 1 to guide optimization efforts. Biochemical assays of ten newly synthesized compounds led to the identification of compound 2, which exhibits a threefold increase in inhibitory activity in vitro relative to 1. In addition, our computational analyses unveiled a surface groove at the interface of the Arp2 and Arp3 subunits that can be exploited for additional structure-based optimization.
Organizational Affiliation:
Department of Chemistry, University of Oregon, Eugene, OR 97403-1253, USA.