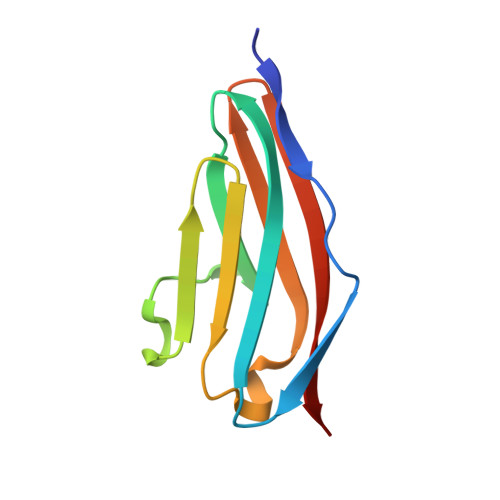
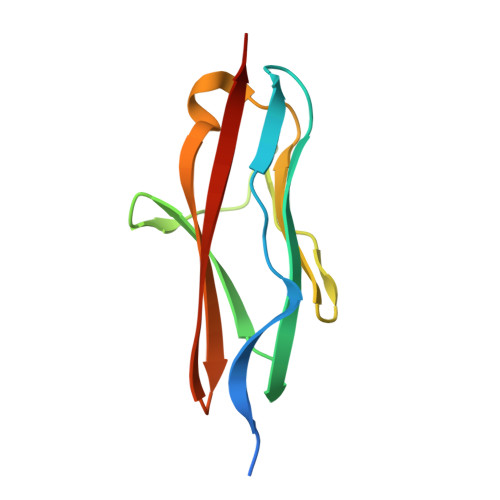
Molecular basis of the head-to-tail assembly of giant muscle proteins obscurin-like 1 and titin.
Sauer, F., Vahokoski, J., Song, Y.H., Wilmanns, M.(2010) EMBO Rep 11: 534-540
- PubMed: 20489725
- DOI: https://doi.org/10.1038/embor.2010.65
- Primary Citation of Related Structures:
3KNB - PubMed Abstract:
Large filament proteins in muscle sarcomeres comprise many immunoglobulin-like domains that provide a molecular platform for self-assembly and interactions with heterologous protein partners. We have unravelled the molecular basis for the head-to-tail interaction of the carboxyl terminus of titin and the amino-terminus of obscurin-like-1 by X-ray crystallography. The binary complex is formed by a parallel intermolecular beta-sheet that presents a novel immunoglobulin-like domain-mediated assembly mechanism in muscle filament proteins. Complementary binding data show that the assembly is entropy-driven rather than dominated data by specific polar interactions. The assembly observed leads to a V-shaped zipper-like arrangement of the two filament proteins.
Organizational Affiliation:
EMBL-Hamburg, Notkestrasse 85, Hamburg D-22603, Germany.