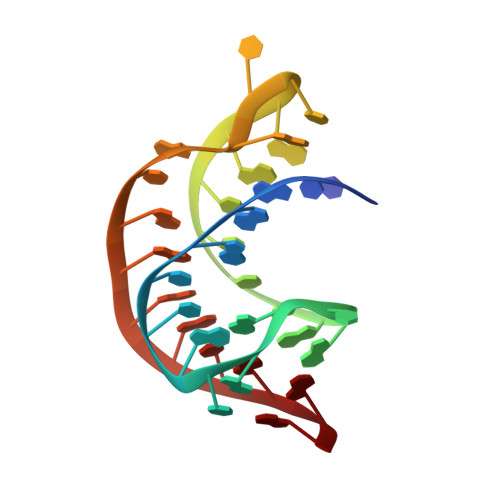
The Structural Basis for Recognition of the PreQ0 Metabolite by an Unusually Small Riboswitch Aptamer Domain.
Spitale, R.C., Torelli, A.T., Krucinska, J., Bandarian, V., Wedekind, J.E.(2009) J Biol Chem 284: 11012-11016
- PubMed: 19261617
- DOI: https://doi.org/10.1074/jbc.C900024200
- Primary Citation of Related Structures:
3GCA - PubMed Abstract:
Riboswitches are RNA elements that control gene expression through metabolite binding. The preQ(1) riboswitch exhibits the smallest known ligand-binding domain and is of interest for its economical organization and high affinity interactions with guanine-derived metabolites required to confer tRNA wobbling. Here we present the crystal structure of a preQ(1) aptamer domain in complex with its precursor metabolite preQ(0). The structure is highly compact with a core that features a stem capped by a well organized decaloop. The metabolite is recognized within a deep pocket via Watson-Crick pairing with C15. Additional hydrogen bonds are made to invariant bases U6 and A29. The ligand-bound state confers continuous helical stacking throughout the core fold, thus providing a platform to promote Watson-Crick base pairing between C9 of the decaloop and the first base of the ribosome-binding site, G33. The structure offers insight into the mode of ribosome-binding site sequestration by a minimal RNA fold stabilized by metabolite binding and has implications for understanding the molecular basis by which bacterial genes are regulated.
Organizational Affiliation:
Department of Biochemistry and Biophysics, University of Rochester School of Medicine and Dentistry, Rochester, New York 14642, USA.