Structural characterization of the protein cce_0567 from Cyanothece 51142, a metalloprotein associated with nitrogen fixation in the DUF683 family.
Buchko, G.W., Robinson, H., Addlagatta, A.(2009) Biochim Biophys Acta 1794: 627-633
- PubMed: 19336042
- DOI: https://doi.org/10.1016/j.bbapap.2009.01.002
- Primary Citation of Related Structures:
3CSX - PubMed Abstract:
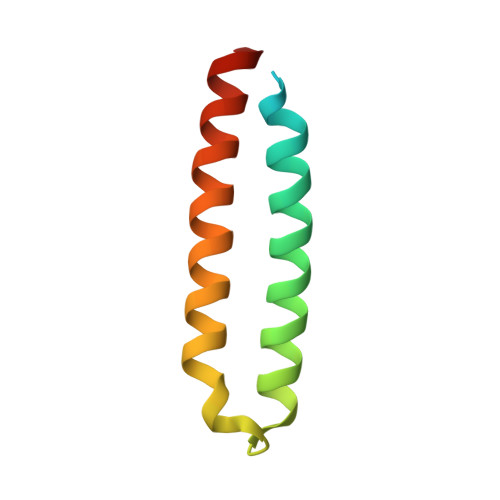
The genomes of many cyanobacteria contain the sequence for a small protein with a common "Domain of Unknown Function" grouped into the DUF683 protein family. While the biological function of DUF683 is still not known, their genomic location within nitrogen fixation clusters suggests that DUF683 proteins may play a role in the process. The diurnal cyanobacterium Cyanothece sp. PCC 51142 contains a gene for a protein that falls into the DUF683 family, cce_0567 (78 aa, 9.0 kDa). In an effort to elucidate the biochemical role DUF683 proteins may play in nitrogen fixation, we have determined the first crystal structure for a protein in this family, cce_0567, to 1.84 A resolution. Cce_0567 crystallized in space group P2(1) with two protein molecules and one Ni(2+) cation per asymmetric unit. The protein is composed of two alpha-helices, residues P11 to G41 (alpha1) and L49-E74 (alpha2), with the second alpha-helix containing a short 3(10)-helix (Y46-N48). A four-residue linker (L42-D45) between the helices allows them to form an anti-parallel bundle and cross over each other towards their termini. In solution it is likely that two molecules of cce_0567 form a rod-like dimer by the stacking interactions of approximately 1/2 of the protein. Histidine-36 is highly conserved in all known DUF683 proteins and the N2 nitrogen of the H36 side chain of each molecule in the dimer is coordinated with Ni(2+) in the crystal structure. The divalent cation Ni(2+) was titrated into (15)N-labeled cce_0567 and chemical shift perturbations were observed only in the (1)H-(15)N HSQC spectra for residues at, or near, the site of Ni(2+) binding observed in the crystal structure. There was no evidence for an increase in the size of cce_0567 upon binding Ni(2+), even in large molar excess of Ni(2+), indicating that a metal was not required for dimer formation. Circular dichroism spectroscopy indicated that cce_0567 was extremely robust, with a melting temperature of approximately 62 degrees C that was reversible.
Organizational Affiliation:
Biological Sciences Division, Pacific Northwest National Laboratory, Richland, WA 99352, USA. garry.buchko@pnl.gov