Holo-Ni(2+)Helicobacter Pylori Nikr Contains Four Square-Planar Nickel-Binding Sites at Physiological Ph.
Benini, S., Cianci, M., Ciurli, S.(2011) Dalton Trans 40: 7831
- PubMed: 21725560
- DOI: https://doi.org/10.1039/c1dt11107h
- Primary Citation of Related Structures:
2Y3Y - PubMed Abstract:
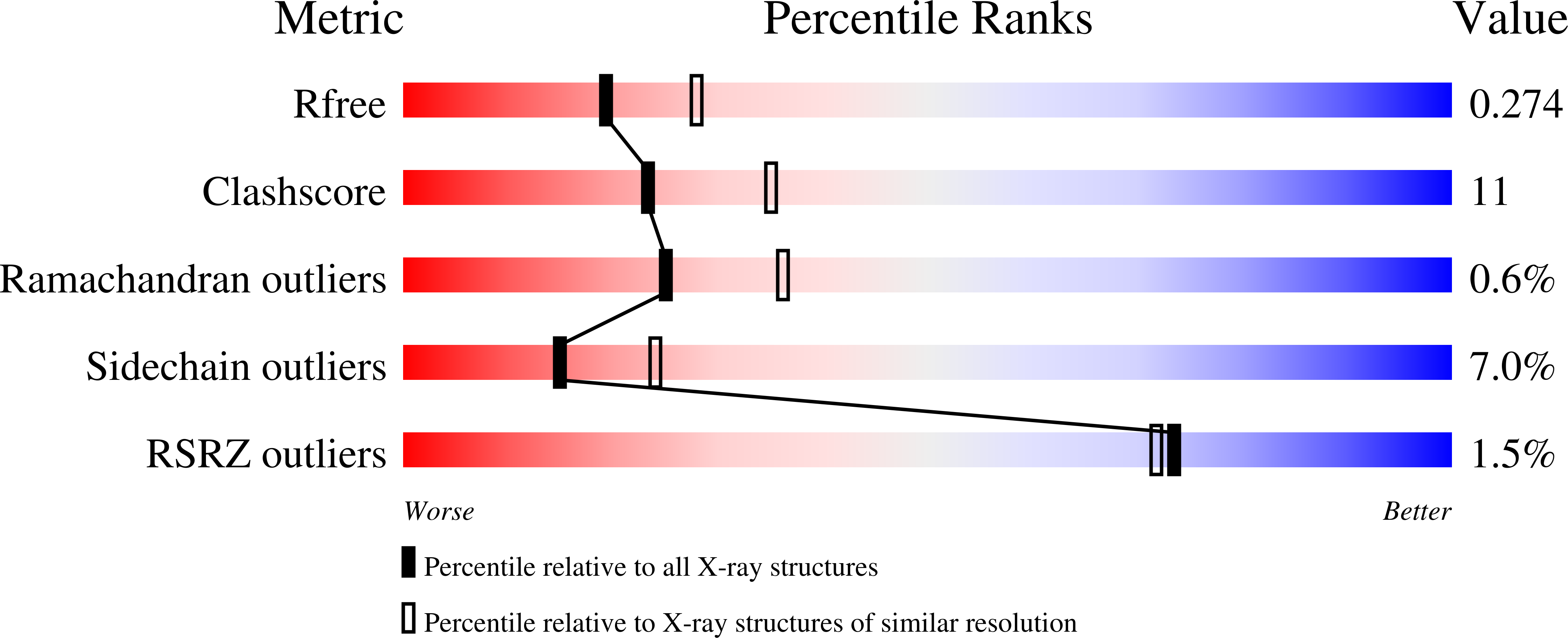
The crystal structure of Helicobacter pylori holo-NikR, a Ni(2+)-dependent transcription factor, determined at pH 7.3, shows four square-planar nickel-binding sites, involving one cysteinate and three histidine ligands. This observation reconciles previous inconsistencies among calorimetric data, structural information at non-physiological pH, and computational studies.
Organizational Affiliation:
Faculty of Science and Technology, Free University of Bolzano, 39100, Bolzano, Italy. stefano.benini@unibz.it