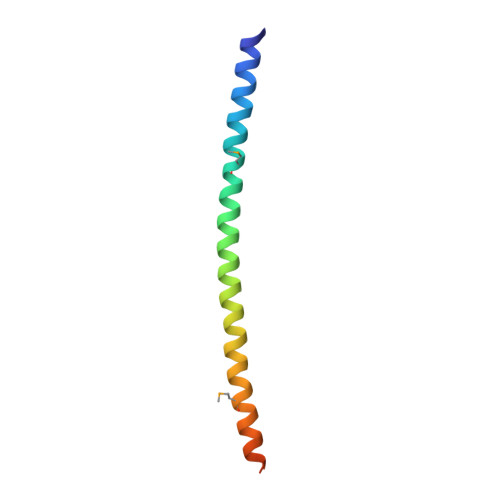Molecular Architecture of a Dynamin Adaptor: Implications for Assembly of Mitochondrial Fission Complexes
Koirala, S., Bui, H.T., Schubert, H.L., Eckert, D.M., Hill, C.P., Kay, M.S., Shaw, J.M.(2010) J Cell Biol 191: 1127
- PubMed: 21149566
- DOI: https://doi.org/10.1083/jcb.201005046
- Primary Citation of Related Structures:
2XU6 - PubMed Abstract:
Recruitment and assembly of some dynamin-related guanosine triphosphatases depends on adaptor proteins restricted to distinct cellular membranes. The yeast Mdv1 adaptor localizes to mitochondria by binding to the membrane protein Fis1. Subsequent Mdv1 binding to the mitochondrial dynamin Dnm1 stimulates Dnm1 assembly into spirals, which encircle and divide the mitochondrial compartment. In this study, we report that dimeric Mdv1 is joined at its center by a 92-Å antiparallel coiled coil (CC). Modeling of the Fis1-Mdv1 complex using available crystal structures suggests that the Mdv1 CC lies parallel to the bilayer with N termini at opposite ends bound to Fis1 and C-terminal β-propeller domains (Dnm1-binding sites) extending into the cytoplasm. A CC length of appropriate length and sequence is necessary for optimal Mdv1 interaction with Fis1 and Dnm1 and is important for proper Dnm1 assembly before membrane scission. Our results provide a framework for understanding how adaptors act as scaffolds to orient and stabilize the assembly of dynamins on membranes.
Organizational Affiliation:
Department of Biochemistry, University of Utah, Salt Lake City, UT 84112, USA.