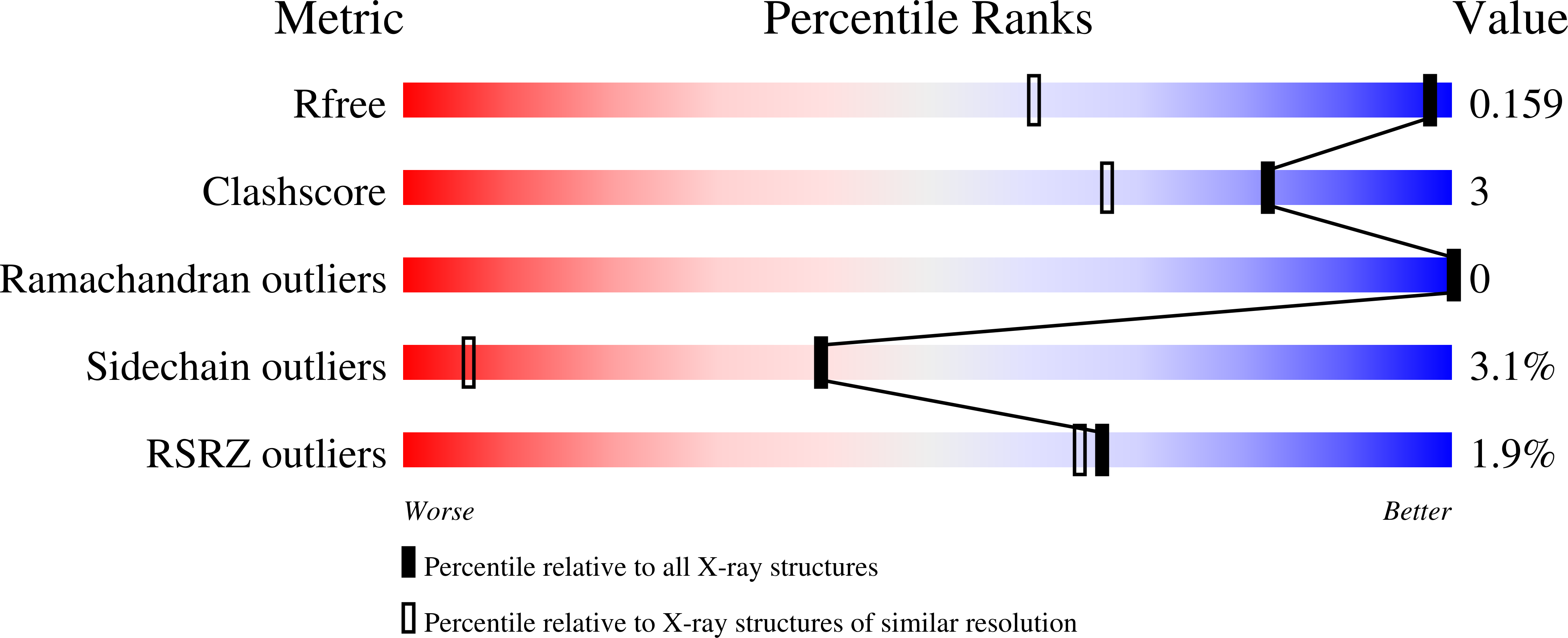
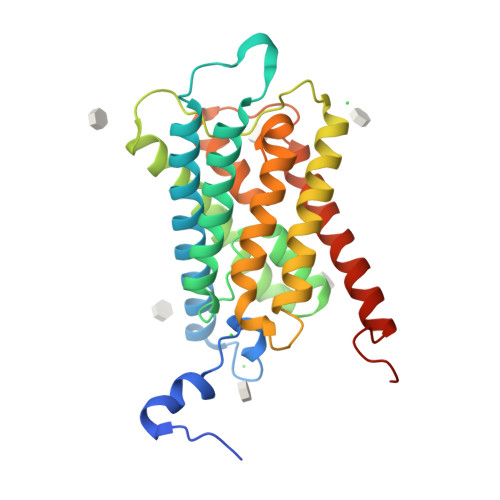
Crystal Structure of a Yeast Aquaporin at 1.15 A Reveals a Novel Gating Mechanism.1.15 A
Fischer, G., Kosinska-Eriksson, U., Aponte-Santamaria, C., Palmgren, M., Geijer, C., Hedfalk, K., Hohmann, S., De Groot, B.L., Neutze, R., Lindkvist-Petersson, K.(2009) PLoS Biol 7: E130
- PubMed: 19529756
- DOI: https://doi.org/10.1371/journal.pbio.1000130
- Primary Citation of Related Structures:
2W1P, 2W2E - PubMed Abstract:
Aquaporins are transmembrane proteins that facilitate the flow of water through cellular membranes. An unusual characteristic of yeast aquaporins is that they frequently contain an extended N terminus of unknown function. Here we present the X-ray structure of the yeast aquaporin Aqy1 from Pichia pastoris at 1.15 A resolution. Our crystal structure reveals that the water channel is closed by the N terminus, which arranges as a tightly wound helical bundle, with Tyr31 forming H-bond interactions to a water molecule within the pore and thereby occluding the channel entrance. Nevertheless, functional assays show that Aqy1 has appreciable water transport activity that aids survival during rapid freezing of P. pastoris. These findings establish that Aqy1 is a gated water channel. Mutational studies in combination with molecular dynamics simulations imply that gating may be regulated by a combination of phosphorylation and mechanosensitivity.
Organizational Affiliation:
Department of Chemistry, Biochemistry and Biophysics, University of Gothenburg, Göteborg, Sweden.