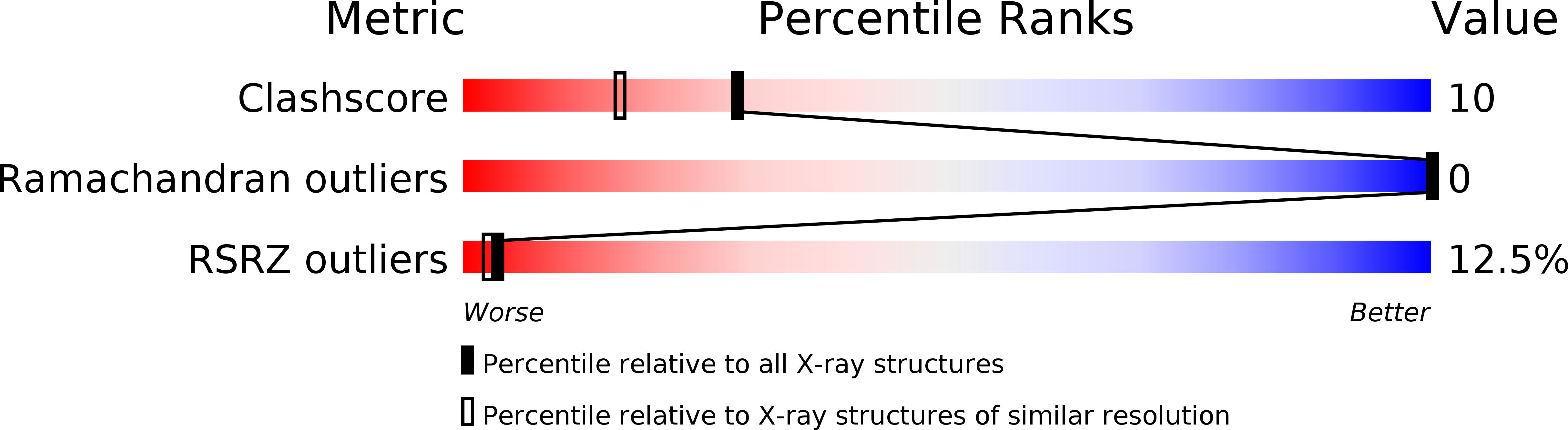Structure of an Enantiomeric Protein, D-Monellin at 1.8 A Resolution.
Hung, L.-W., Kohmura, M., Ariyoshi, Y., Kim, S.-H.(1998) Acta Crystallogr D Biol Crystallogr 54: 494-500
- PubMed: 9867435
- DOI: https://doi.org/10.1107/s0907444997012225
- Primary Citation of Related Structures:
2Q33 - PubMed Abstract:
The D-enantiomer of a potently sweet protein, monellin, has been crystallized and analyzed by X-ray crystallography at 1.8 A resolut ion. Two crystal forms (I and II) appeared under crystallization conditions similar, but not identical, to the crystallization conditions of natural L-monellin. There are four molecules per asymmetric unit in crystal form I and one in crystal form II. Crystal form I is not reproducible and is equivalent to that of monoclinic L-monellin. Intermonomer contacts in crystal form II are very different from those found in natural L-monellin crystals. The backbone trace of D-monellin resembles very closely the mirror image of that of L-monellin, but the N- and C-terminus backbones as well as several side-chain conformations of D-monellin are different from those of natural L-monellin. Most of these apparent differences may be attributable to the crystal packing differences.
Organizational Affiliation:
Graduate Group in Biophysics, Department of Chemistry, Univesity of California, Berkeley, 94720, USA.