Structure of CD84 provides insight into SLAM family function.
Yan, Q., Malashkevich, V.N., Fedorov, A., Fedorov, E., Cao, E., Lary, J.W., Cole, J.L., Nathenson, S.G., Almo, S.C.(2007) Proc Natl Acad Sci U S A 104: 10583-10588
- PubMed: 17563375
- DOI: https://doi.org/10.1073/pnas.0703893104
- Primary Citation of Related Structures:
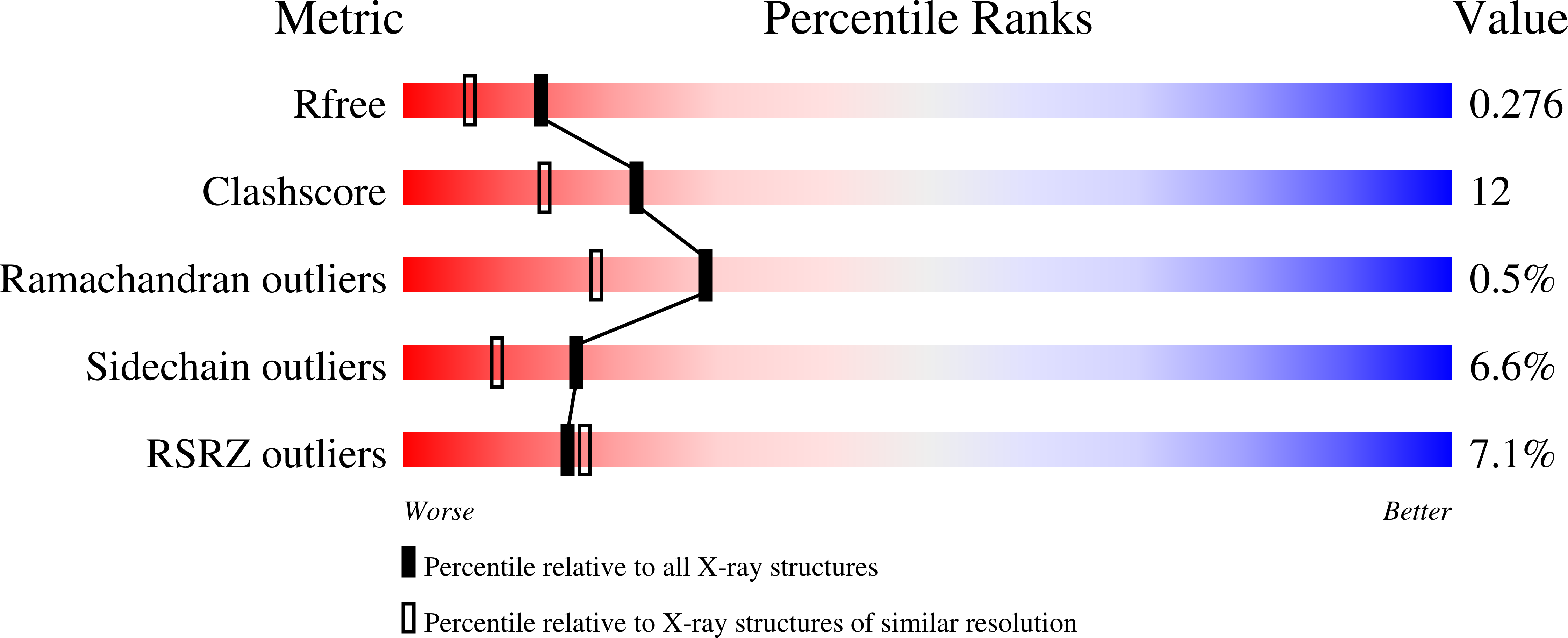
2PKD - PubMed Abstract:
The signaling lymphocyte activation molecule (SLAM) family includes homophilic and heterophilic receptors that modulate both adaptive and innate immune responses. These receptors share a common ectodomain organization: a membrane-proximal immunoglobulin constant domain and a membrane-distal immunoglobulin variable domain that is responsible for ligand recognition. CD84 is a homophilic family member that enhances IFN-gamma secretion in activated T cells. Our solution studies revealed that CD84 strongly self-associates with a K(d) in the submicromolar range. These data, in combination with previous reports, demonstrate that the SLAM family homophilic affinities span at least three orders of magnitude and suggest that differences in the affinities may contribute to the distinct signaling behavior exhibited by the individual family members. The 2.0 A crystal structure of the human CD84 immunoglobulin variable domain revealed an orthogonal homophilic dimer with high similarity to the recently reported homophilic dimer of the SLAM family member NTB-A. Structural and chemical differences in the homophilic interfaces provide a mechanism to prevent the formation of undesired heterodimers among the SLAM family homophilic receptors. These structural data also suggest that, like NTB-A, all SLAM family homophilic dimers adopt a highly kinked organization spanning an end-to-end distance of approximately 140 A. This common molecular dimension provides an opportunity for all two-domain SLAM family receptors to colocalize within the immunological synapse and bridge the T cell and antigen-presenting cell.
Organizational Affiliation:
Department of Cell Biology, Albert Einstein College of Medicine, Bronx, NY 10461, USA.