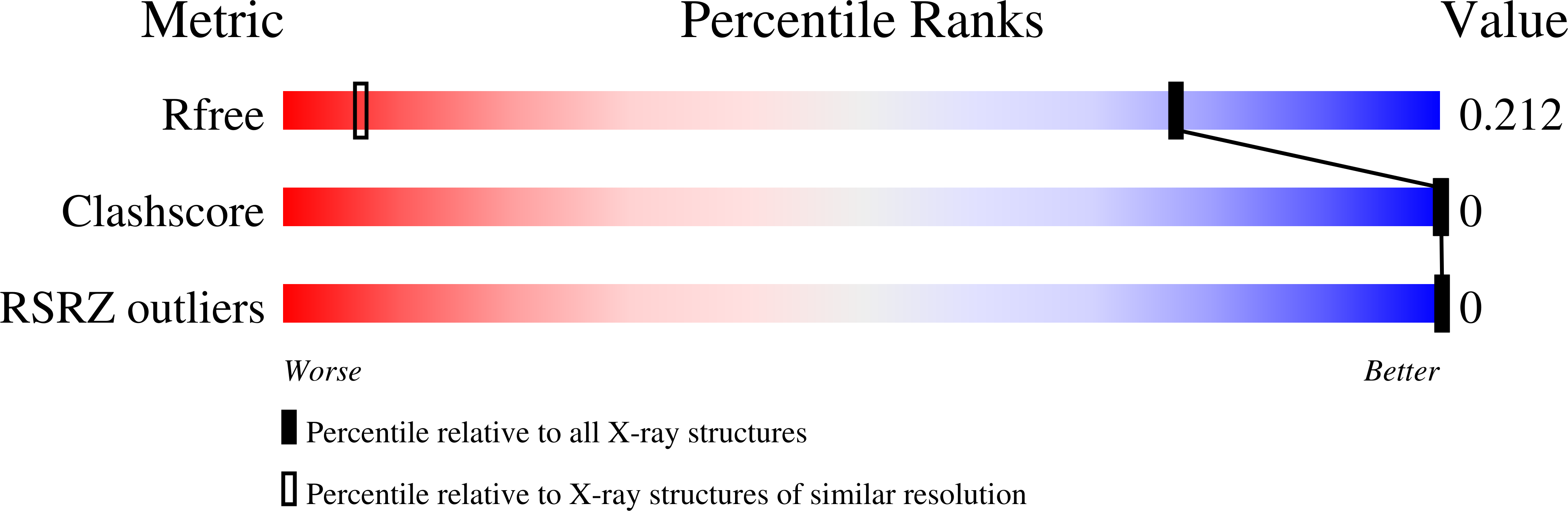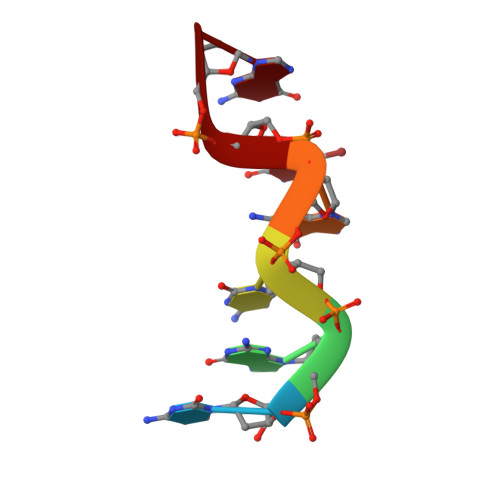Polarisation-dependence of anomalous scattering in brominated DNA and RNA molecules, and importance of crystal orientation in SAD and MAD phasing
Sanishvili, R., Besnard, C., Camus, F., Fleurant, M., Pattison, P., Bricongne, G., Schiltz, M.To be published.