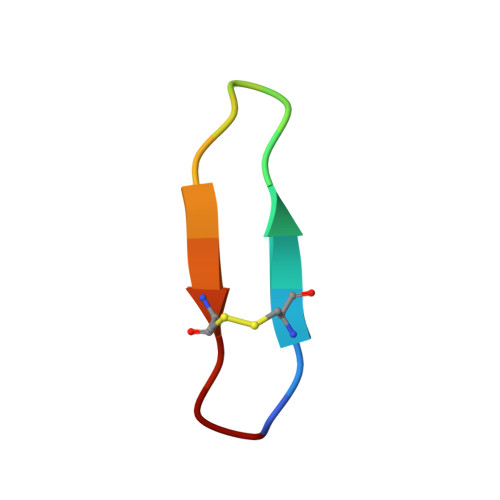
Diverse cyclic seed peptides in the Mexican zinnia (Zinnia haageana).
Franke, B., Jayasena, A.S., Fisher, M.F., Swedberg, J.E., Taylor, N.L., Mylne, J.S., Rosengren, K.J.(2016) Biopolymers 106: 806-817
- PubMed: 27352920
- DOI: https://doi.org/10.1002/bip.22901
- Primary Citation of Related Structures:
2NDL, 2NDM, 2NDN - PubMed Abstract:
A new family of small plant peptides was recently described and found to be widespread throughout the Millereae and Heliantheae tribes of the sunflower family Asteraceae. These peptides originate from the post-translational processing of unusual seed-storage albumin genes, and have been termed PawS-derived peptides (PDPs). The prototypic family member is a 14-residue cyclic peptide with potent trypsin inhibitory activity named SunFlower Trypsin Inhibitor (SFTI-1). In this study we present the features of three new PDPs discovered in the seeds of the sunflower species Zinnia haageana by a combination of de novo transcriptomics and liquid chromatography-mass spectrometry. Two-dimensional solution NMR spectroscopy was used to elucidate their structural characteristics. All three Z. haageana peptides have well-defined folds with a head-to-tail cyclized peptide backbone and a single disulfide bond. Although two possess an anti-parallel β-sheet structure, like SFTI-1, the Z. haageana peptide PDP-21 has a more irregular backbone structure. Despite structural similarities with SFTI-1, PDP-20 was not able to inhibit trypsin, thus the functional roles of these peptides is yet to be discovered. Defining the structural features of the small cyclic peptides found in the sunflower family will be useful for guiding the exploitation of these peptides as scaffolds for grafting and protein engineering applications.
Organizational Affiliation:
School of Biomedical Sciences, The University of Queensland, St Lucia, QLD, 4072, Australia.