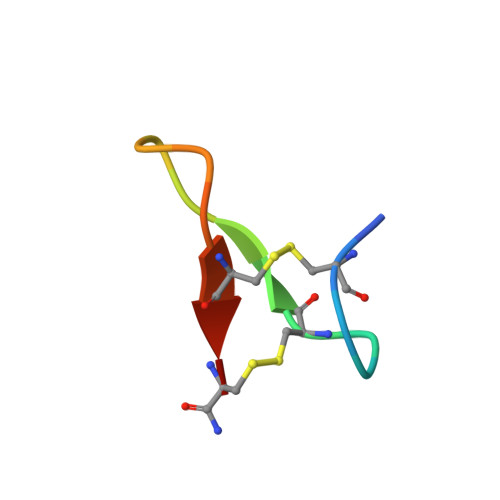
Isolation and characterization of a structurally unique beta-hairpin venom peptide from the predatory ant Anochetus emarginatus.
Touchard, A., Brust, A., Cardoso, F.C., Chin, Y.K., Herzig, V., Jin, A.H., Dejean, A., Alewood, P.F., King, G.F., Orivel, J., Escoubas, P.(2016) Biochim Biophys Acta 1860: 2553-2562
- PubMed: 27474999
- DOI: https://doi.org/10.1016/j.bbagen.2016.07.027
- Primary Citation of Related Structures:
2NBC - PubMed Abstract:
Most ant venoms consist predominantly of small linear peptides, although some contain disulfide-linked peptides as minor components. However, in striking contrast to other ant species, some Anochetus venoms are composed primarily of disulfide-rich peptides. In this study, we investigated the venom of the ant Anochetus emarginatus with the aim of exploring these novel disulfide-rich peptides.
Organizational Affiliation:
CNRS, UMR Ecologie des forêts de Guyane (AgroParisTech, CIRAD, CNRS, INRA, Université de Guyane, Université des Antilles), Campus Agronomique, BP 316, 97379 Kourou, France. Electronic address: t.axel@hotmail.fr.