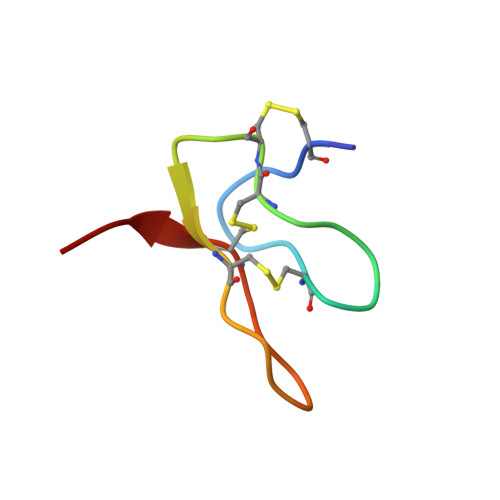
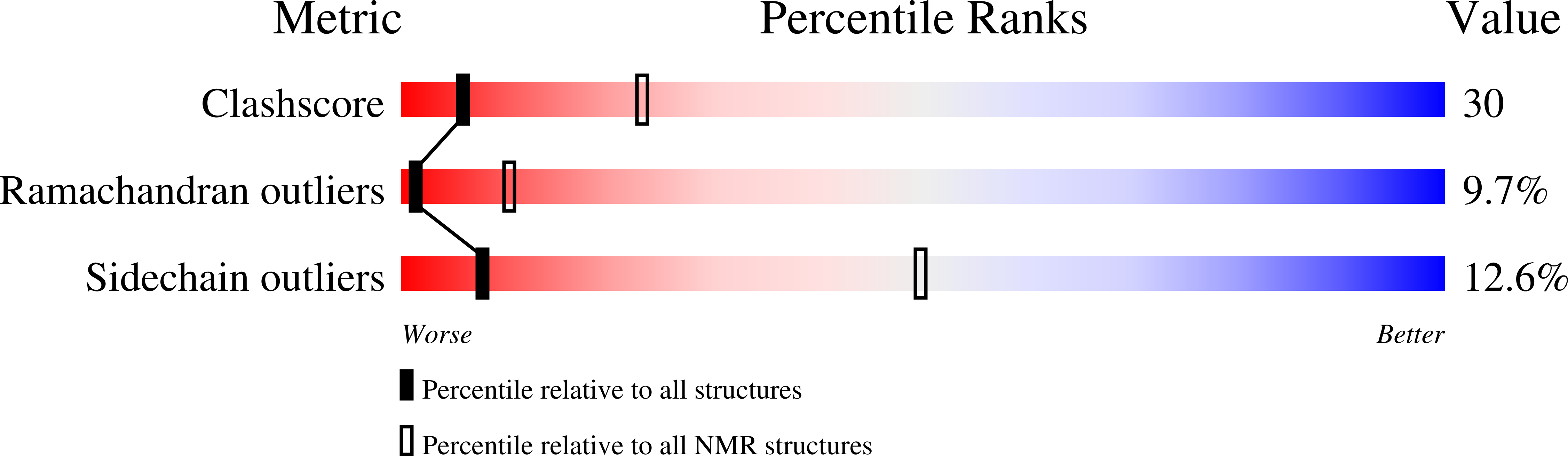Structural insights into the mechanism of activation of the TRPV1 channel by a membrane-bound tarantula toxin
Bae, C., Anselmi, C., Kalia, J., Jara-Oseguera, A., Schwieters, C.D., Krepkiy, D., Lee, C.W., Kim, E.H., Kim, J.I., Faraldo-Gomez, J.D., Swartz, K.J.(2016) Elife 5
- PubMed: 26880553
- DOI: https://doi.org/10.7554/eLife.11273
- Primary Citation of Related Structures:
2N9Z, 2NAJ - PubMed Abstract:
Venom toxins are invaluable tools for exploring the structure and mechanisms of ion channels. Here, we solve the structure of double-knot toxin (DkTx), a tarantula toxin that activates the heat-activated TRPV1 channel. We also provide improved structures of TRPV1 with and without the toxin bound, and investigate the interactions of DkTx with the channel and membranes. We find that DkTx binds to the outer edge of the external pore of TRPV1 in a counterclockwise configuration, using a limited protein-protein interface and inserting hydrophobic residues into the bilayer. We also show that DkTx partitions naturally into membranes, with the two lobes exhibiting opposing energetics for membrane partitioning and channel activation. Finally, we find that the toxin disrupts a cluster of hydrophobic residues behind the selectivity filter that are critical for channel activation. Collectively, our findings reveal a novel mode of toxin-channel recognition that has important implications for the mechanism of thermosensation.
Organizational Affiliation:
Molecular Physiology and Biophysics Section, Porter Neuroscience Research Center, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, United States.