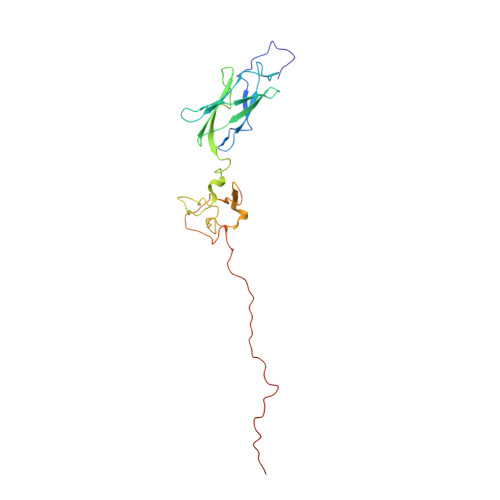
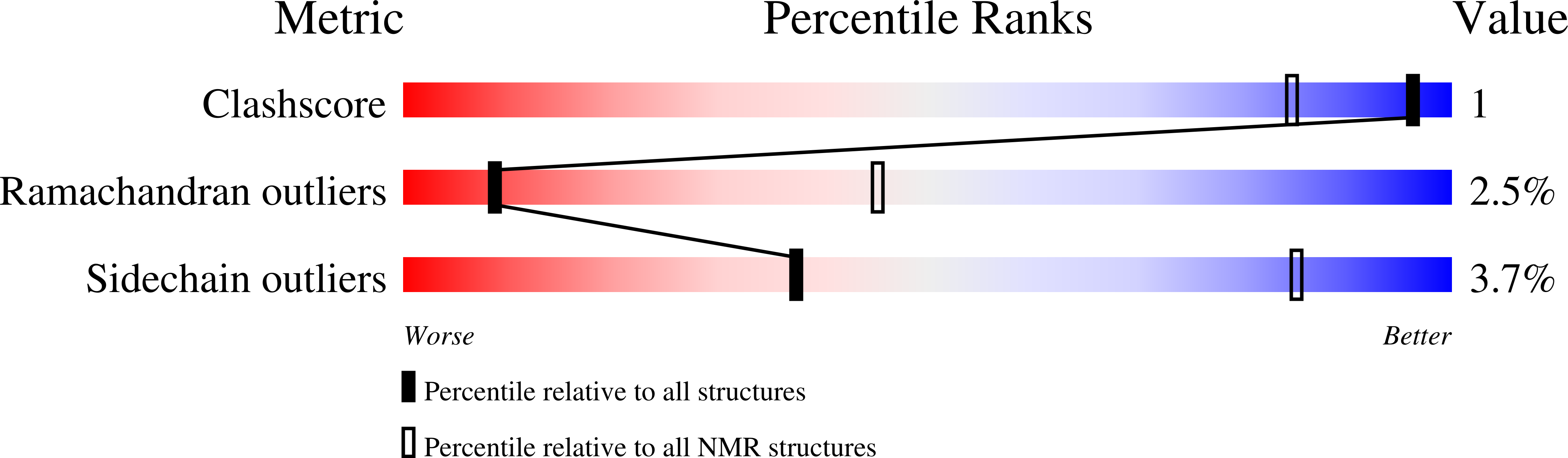A structural ensemble of a ribosome-nascent chain complex during cotranslational protein folding.
Cabrita, L.D., Cassaignau, A.M., Launay, H.M., Waudby, C.A., Wlodarski, T., Camilloni, C., Karyadi, M.E., Robertson, A.L., Wang, X., Wentink, A.S., Goodsell, L.S., Woolhead, C.A., Vendruscolo, M., Dobson, C.M., Christodoulou, J.(2016) Nat Struct Mol Biol 23: 278-285
- PubMed: 26926436
- DOI: https://doi.org/10.1038/nsmb.3182
- Primary Citation of Related Structures:
2N62 - PubMed Abstract:
Although detailed pictures of ribosome structures are emerging, little is known about the structural and cotranslational folding properties of nascent polypeptide chains at the atomic level. Here we used solution-state NMR spectroscopy to define a structural ensemble of a ribosome-nascent chain complex (RNC) formed during protein biosynthesis in Escherichia coli, in which a pair of immunoglobulin-like domains adopts a folded N-terminal domain (FLN5) and a disordered but compact C-terminal domain (FLN6). To study how FLN5 acquires its native structure cotranslationally, we progressively shortened the RNC constructs. We found that the ribosome modulates the folding process, because the complete sequence of FLN5 emerged well beyond the tunnel before acquiring native structure, whereas FLN5 in isolation folded spontaneously, even when truncated. This finding suggests that regulating structure acquisition during biosynthesis can reduce the probability of misfolding, particularly of homologous domains.
Organizational Affiliation:
institute of Structural and Molecular Biology, University College London.