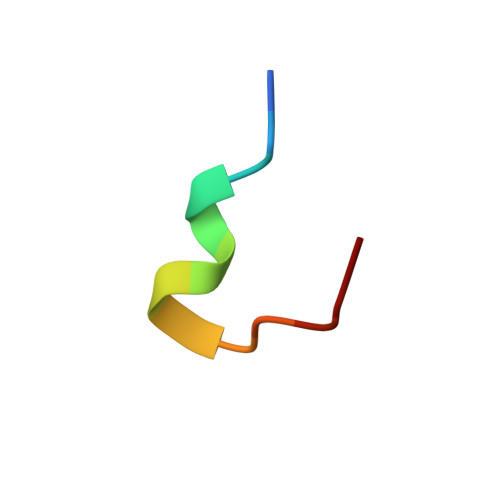
NMR structure and dynamics of the agonist dynorphin peptide bound to the human kappa opioid receptor.
O'Connor, C., White, K.L., Doncescu, N., Didenko, T., Roth, B.L., Czaplicki, G., Stevens, R.C., Wuthrich, K., Milon, A.(2015) Proc Natl Acad Sci U S A 112: 11852-11857
- PubMed: 26372966
- DOI: https://doi.org/10.1073/pnas.1510117112
- Primary Citation of Related Structures:
2N2F - PubMed Abstract:
The structure of the dynorphin (1-13) peptide (dynorphin) bound to the human kappa opioid receptor (KOR) has been determined by liquid-state NMR spectroscopy. (1)H and (15)N chemical shift variations indicated that free and bound peptide is in fast exchange in solutions containing 1 mM dynorphin and 0.01 mM KOR. Radioligand binding indicated an intermediate-affinity interaction, with a Kd of ∼200 nM. Transferred nuclear Overhauser enhancement spectroscopy was used to determine the structure of bound dynorphin. The N-terminal opioid signature, YGGF, was observed to be flexibly disordered, the central part of the peptide from L5 to R9 to form a helical turn, and the C-terminal segment from P10 to K13 to be flexibly disordered in this intermediate-affinity bound state. Combining molecular modeling with NMR provided an initial framework for understanding multistep activation of a G protein-coupled receptor by its cognate peptide ligand.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037; Departments of Biological Sciences and Chemistry, Bridge Institute, University of Southern California, Los Angeles, CA 90089;
Organizational Affiliation: