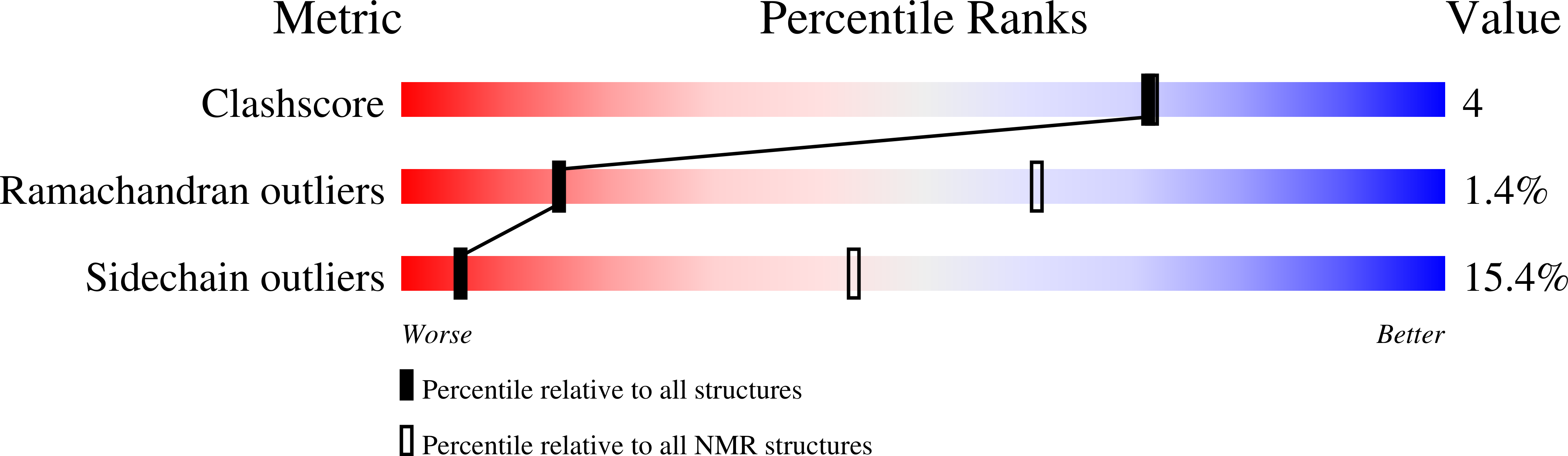Structure-based drug design identifies polythiophenes as antiprion compounds.
Herrmann, U.S., Schutz, A.K., Shirani, H., Huang, D., Saban, D., Nuvolone, M., Li, B., Ballmer, B., Aslund, A.K., Mason, J.J., Rushing, E., Budka, H., Nystrom, S., Hammarstrom, P., Bockmann, A., Caflisch, A., Meier, B.H., Nilsson, K.P., Hornemann, S., Aguzzi, A.(2015) Sci Transl Med 7: 299ra123-299ra123
- PubMed: 26246168
- DOI: https://doi.org/10.1126/scitranslmed.aab1923
- Primary Citation of Related Structures:
2MUS - PubMed Abstract:
Prions cause transmissible spongiform encephalopathies for which no treatment exists. Prions consist of PrP(Sc), a misfolded and aggregated form of the cellular prion protein (PrP(C)). We explore the antiprion properties of luminescent conjugated polythiophenes (LCPs) that bind and stabilize ordered protein aggregates. By administering a library of structurally diverse LCPs to the brains of prion-infected mice via osmotic minipumps, we found that antiprion activity required a minimum of five thiophene rings bearing regularly spaced carboxyl side groups. Solid-state nuclear magnetic resonance analyses and molecular dynamics simulations revealed that anionic side chains interacted with complementary, regularly spaced cationic amyloid residues of model prions. These findings allowed us to extract structural rules governing the interaction between LCPs and protein aggregates, which we then used to design a new set of LCPs with optimized binding. The new set of LCPs showed robust prophylactic and therapeutic potency in prion-infected mice, with the lead compound extending survival by >80% and showing activity against both mouse and hamster prions as well as efficacy upon intraperitoneal administration into mice. These results demonstrate the feasibility of targeted chemical design of compounds that may be useful for treating diseases of aberrant protein aggregation such as prion disease.
Organizational Affiliation:
Institute of Neuropathology, University Hospital of Zürich, University of Zürich, 8091 Zürich, Switzerland.