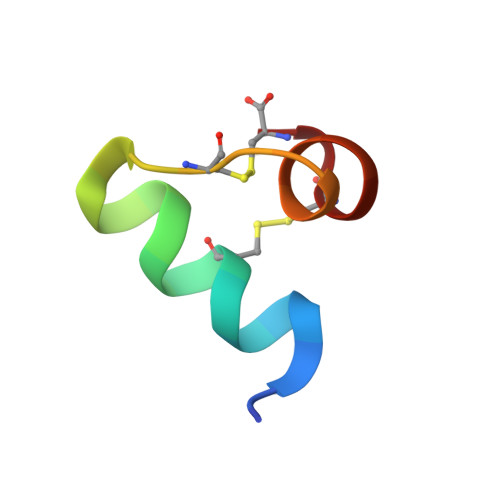
Structural basis for antimicrobial activity of lasiocepsin.
Monincova, L., Budesinsky, M., Cujova, S., Cerovsky, V., Veverka, V.(2014) Chembiochem 15: 301-308
- PubMed: 24339323
- DOI: https://doi.org/10.1002/cbic.201300509
- Primary Citation of Related Structures:
2MBD - PubMed Abstract:
Lasiocepsin is a unique 27-residue antimicrobial peptide, isolated from Lasioglossum laticeps (wild bee) venom, with substantial antibacterial and antifungal activity. It adopts a well-defined structure consisting of two α-helices linked by a structured loop. Its basic residues form two distinct positively charged regions on the surface whereas aliphatic side chains contribute to solvent-accessible hydrophobic areas, thus emphasising the amphipathic character of the molecule. Lasiocepsin structurally belongs to the ShK family and shows a strong preference for anionic phospholipids; this is further augmented by increasing concentrations of cardiolipin, such as those found at the poles of bacterial cells. The membrane-permeabilising activity of the peptide is not limited to outer membranes of Gram-negative bacteria. The peptide interacts with phospholipids initially through its N terminus, and its degree of penetration is strongly dependent on the presence of cardiolipin.
Organizational Affiliation:
Institute of Organic Chemistry and Biochemistry, Academy of Sciences of the Czech Republic, Flemingovo nam. 2, 16610 Prague 6 (Czech Republic).