Solution structure of the PsIAA4 oligomerization domain reveals interaction modes for transcription factors in early auxin response.
Dinesh, D.C., Kovermann, M., Gopalswamy, M., Hellmuth, A., Calderon Villalobos, L.I., Lilie, H., Balbach, J., Abel, S.(2015) Proc Natl Acad Sci U S A 112: 6230-6235
- PubMed: 25918389
- DOI: https://doi.org/10.1073/pnas.1424077112
- Primary Citation of Related Structures:
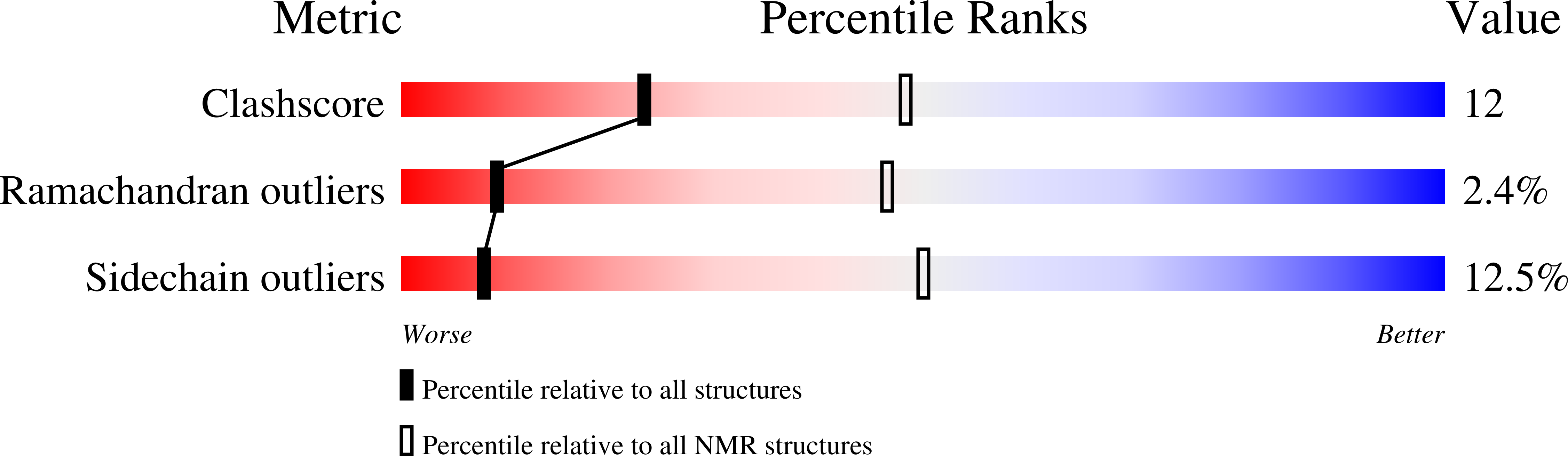
2M1M - PubMed Abstract:
The plant hormone auxin activates primary response genes by facilitating proteolytic removal of auxin/indole-3-acetic acid (AUX/IAA)-inducible repressors, which directly bind to transcriptional auxin response factors (ARF). Most AUX/IAA and ARF proteins share highly conserved C-termini mediating homotypic and heterotypic interactions within and between both protein families. The high-resolution NMR structure of C-terminal domains III and IV of the AUX/IAA protein PsIAA4 from pea (Pisum sativum) revealed a globular ubiquitin-like β-grasp fold with homologies to the Phox and Bem1p (PB1) domain. The PB1 domain of wild-type PsIAA4 features two distinct surface patches of oppositely charged amino acid residues, mediating front-to-back multimerization via electrostatic interactions. Mutations of conserved basic or acidic residues on either face suppressed PsIAA4 PB1 homo-oligomerization in vitro and confirmed directional interaction of full-length PsIAA4 in vivo (yeast two-hybrid system). Mixing of oppositely mutated PsIAA4 PB1 monomers enabled NMR mapping of the negatively charged interface of the reconstituted PsIAA4 PB1 homodimer variant, whose stoichiometry (1:1) and equilibrium binding constant (KD ∼ 6.4 μM) were determined by isothermal titration calorimetry. In silico protein-protein docking studies based on NMR and yeast interaction data derived a model of the PsIAA4 PB1 homodimer, which is comparable with other PB1 domain dimers, but indicated considerable differences between the homodimeric interfaces of AUX/IAA and ARF PB1 domains. Our study provides an impetus for elucidating the molecular determinants that confer specificity to complex protein-protein interaction circuits between members of the two central families of transcription factors important to the regulation of auxin-responsive gene expression.
Organizational Affiliation:
Department of Molecular Signal Processing, Leibniz Institute of Plant Biochemistry, D-06120 Halle, Germany;