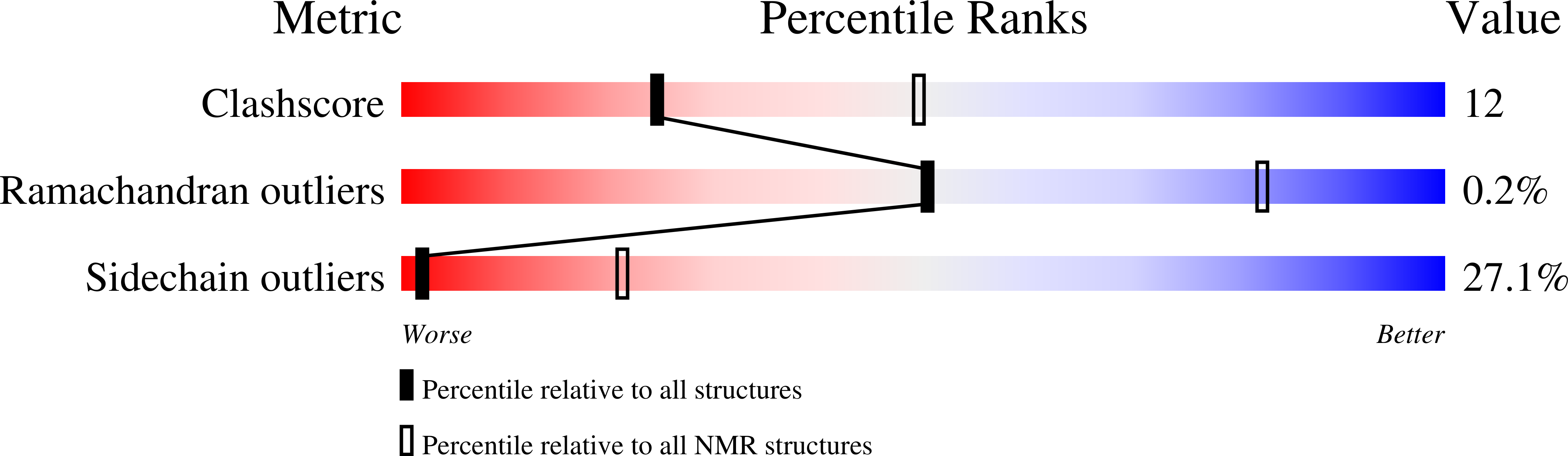The Sam domain of the lipid phosphatase Ship2 adopts a common model to interact with Arap3-Sam and EphA2-Sam.
Leone, M., Cellitti, J., Pellecchia, M.(2009) BMC Struct Biol 9: 59-59
- PubMed: 19765305
- DOI: https://doi.org/10.1186/1472-6807-9-59
- Primary Citation of Related Structures:
2KG5 - PubMed Abstract:
Sterile alpha motif (Sam) domains are small protein modules that can be involved in homotypic or heterotypic associations and exhibit different functions. Previous studies have demonstrated that the Sam domain of the lipid phosphatase Ship2 can hetero-dimerize with the Sam domain of the PI3K effector protein Arap3.
Organizational Affiliation:
Burnham Institute for Medical Research, La Jolla, California, USA. mleone@burnham.org