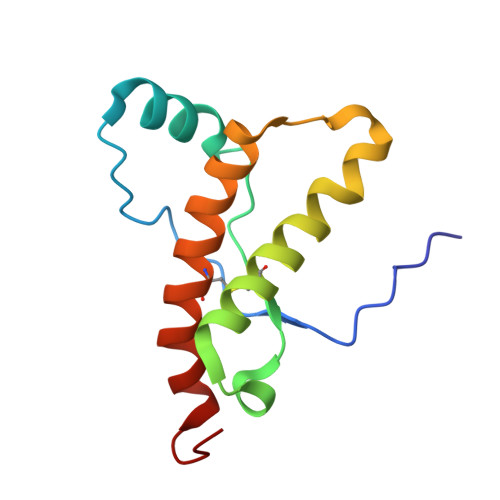
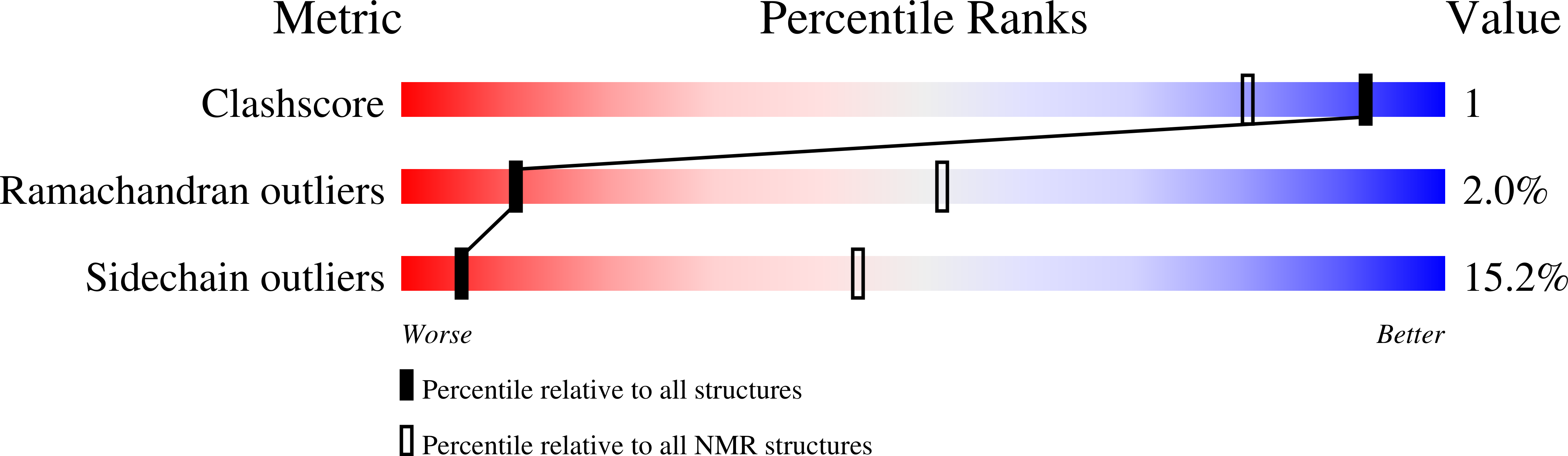NMR Structure of the Bank Vole Prion Protein at 20 degrees C Contains a Structured Loop of Residues 165-171.
Christen, B., Perez, D.R., Hornemann, S., Wuthrich, K.(2008) J Mol Biol 383: 306-312
- PubMed: 18773909
- DOI: https://doi.org/10.1016/j.jmb.2008.08.045
- Primary Citation of Related Structures:
2K56, 2K5O - PubMed Abstract:
The recent introduction of bank vole (Clethrionomys glareolus) as an additional laboratory animal for research on prion diseases revealed an important difference when compared to the mouse and the Syrian hamster, since bank voles show a high susceptibility to infection by brain homogenates from a wide range of diseased species such as sheep, goats, and humans. In this context, we determined the NMR structure of the C-terminal globular domain of the recombinant bank vole prion protein (bvPrP) [bvPrP(121-231)] at 20 degrees C. bvPrP(121-231) has the same overall architecture as other mammalian PrPs, with three alpha-helices and an antiparallel beta-sheet, but it differs from PrP of the mouse and most other mammalian species in that the loop connecting the second beta-strand and helix alpha2 is precisely defined at 20 degrees C. This is similar to the previously described structures of elk PrP and the designed mouse PrP (mPrP) variant mPrP[S170N,N174T](121-231), whereas Syrian hamster PrP displays a structure that is in-between these limiting cases. Studies with the newly designed variant mPrP[S170N](121-231), which contains the same loop sequence as bvPrP, now also showed that the single-amino-acid substitution S170N in mPrP is sufficient for obtaining a well-defined loop, thus providing the rationale for this local structural feature in bvPrP.
Organizational Affiliation:
Institute of Molecular Biology and Biophysics, Schafmattstrasse 20, ETH Zurich, CH-8093 Zurich, Switzerland.