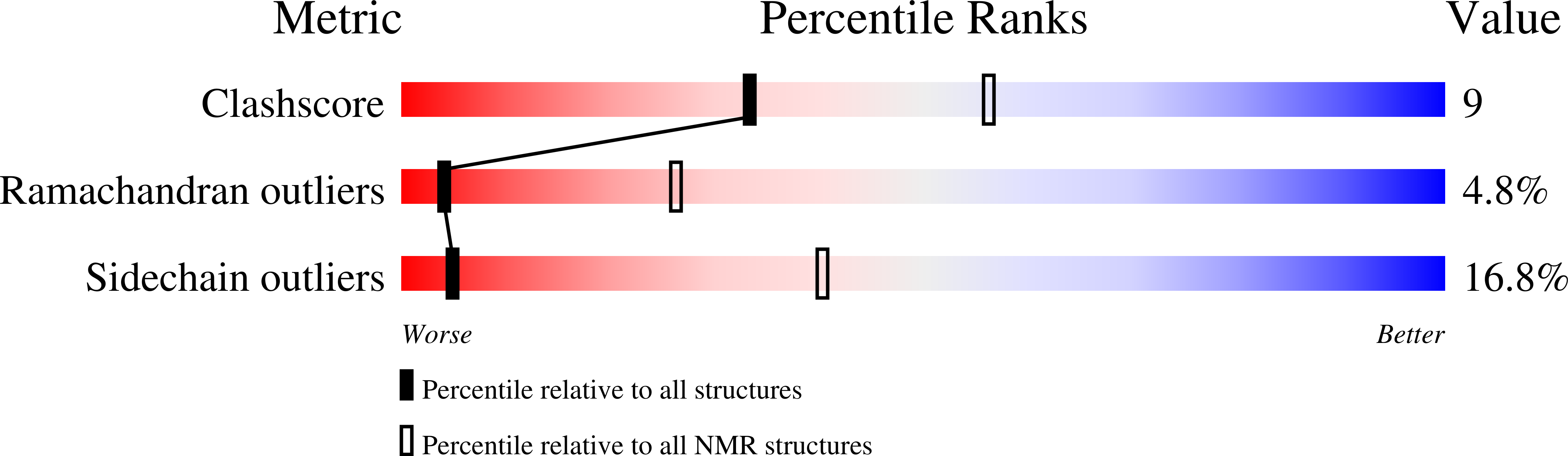Hydramacin-1, structure and antibacterial activity of a protein from the Basal metazoan hydra.
Jung, S., Dingley, A.J., Augustin, R., Anton-Erxleben, F., Stanisak, M., Gelhaus, C., Gutsmann, T., Hammer, M.U., Podschun, R., Bonvin, A.M., Leippe, M., Bosch, T.C., Grotzinger, J.(2009) J Biol Chem 284: 1896-1905
- PubMed: 19019828
- DOI: https://doi.org/10.1074/jbc.M804713200
- Primary Citation of Related Structures:
2K35 - PubMed Abstract:
Hydramacin-1 is a novel antimicrobial protein recently discovered during investigations of the epithelial defense of the ancient metazoan Hydra. The amino acid sequence of hydramacin-1 shows no sequence homology to any known antimicrobial proteins. Determination of the solution structure revealed that hydramacin-1 possesses a disulfide bridge-stabilized alphabeta motif. This motif is the common scaffold of the knottin protein fold. The structurally closest relatives are the scorpion oxin-like superfamily. Within this superfamily hydramacin-1 establishes a new family of proteins that all share antimicrobial activity. Hydramacin-1 is potently active against Gram-positive and Gram-negative bacteria including multi-resistant human pathogenic strains. It leads to aggregation of bacteria as an initial step of its bactericidal mechanism. Aggregated cells are connected via electron-dense contacts and adopt a thorn apple-like morphology. Analysis of the hydramacin-1 structure revealed an unusual distribution of amino acid side chains on the surface. A belt of positively charged residues is sandwiched by two hydrophobic areas. Based on this characteristic surface feature and on biophysical analysis of protein-membrane interactions, we propose a model that describes the aggregation effect exhibited by hydramacin-1.
Organizational Affiliation:
Institute of Biochemistry and Institute of Zoology, Zoophysiology, Christian-Albrechts-University of Kiel, Olshausenstrasse 40, 24098 Kiel, Germany.