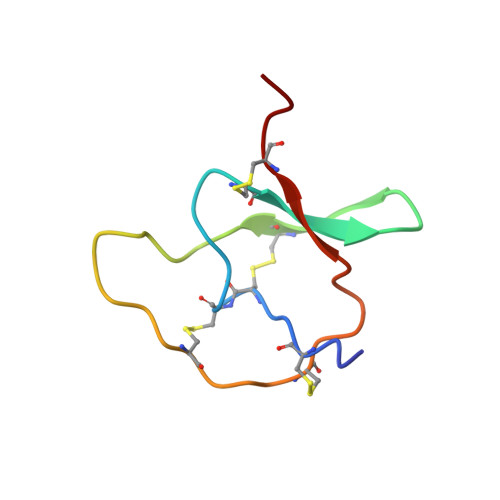
Structural refinement of insecticidal plant proteinase inhibitors from Nicotiana alata.
Schirra, H.J., Anderson, M.A., Craik, D.J.(2008) Protein Pept Lett 15: 903-909
- PubMed: 18991765
- DOI: https://doi.org/10.2174/092986608785849326
- Primary Citation of Related Structures:
2JZM - PubMed Abstract:
Ornamental tobacco (Nicotiana alata) produces a series of 6 kDa proteinase inhibitors belonging to the potato type II inhibitor family. These proteins inhibit trypsin and chymotrypsin, the main digestive enzymes of predatory insects, thus leading to starvation, impaired larval development or death. In this context, the three-dimensional structures of these inhibitors are important for developing novel strategies for pest control. The solution structures of C1 and T1, the two main prototypes of the N. alata inhibitors, were originally determined more than a decade ago (J. Mol. Biol. 242, 231-243 (1994) and Biochemistry 34, 14304-14311 (1995)). Since then methods for NMR structure calculations have evolved considerably. Here we report the refinement of the structures of C1 and T1 with state-of-the-art protocols for NMR structure calculations. This refinement leads to an improved quality of the structures, making them a more reliable basis for the development of novel pesticides and modeling applications.
Organizational Affiliation:
Institute for Molecular Bioscience, The University of Queensland, Brisbane, Qld 4072, Australia.