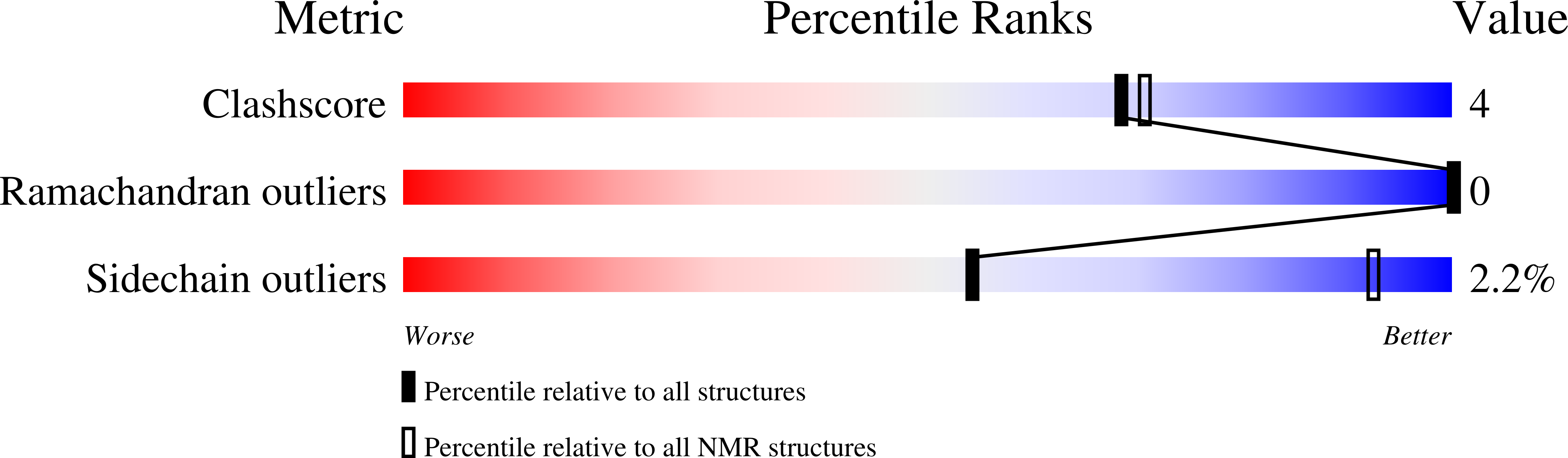Structural diversity in twin-arginine signal peptide-binding proteins.
Maillard, J., Spronk, C.A.E.M., Buchanan, G., Lyall, V., Richardson, D.J., Palmer, T., Vuister, G.W., Sargent, F.(2007) Proc Natl Acad Sci U S A 104: 15641-15646
- PubMed: 17901208
- DOI: https://doi.org/10.1073/pnas.0703967104
- Primary Citation of Related Structures:
2JSX - PubMed Abstract:
The twin-arginine transport (Tat) system is dedicated to the translocation of folded proteins across the bacterial cytoplasmic membrane. Proteins are targeted to the Tat system by signal peptides containing a twin-arginine motif. In Escherichia coli, many Tat substrates bind redox-active cofactors in the cytoplasm before transport. Coordination of cofactor insertion with protein export involves a "Tat proofreading" process in which chaperones bind twin-arginine signal peptides, thus preventing premature export. The initial Tat signal-binding proteins described belonged to the TorD family, which are required for assembly of N- and S-oxide reductases. Here, we report that E. coli NapD is a Tat signal peptide-binding chaperone involved in biosynthesis of the Tat-dependent nitrate reductase NapA. NapD binds tightly and specifically to the NapA twin-arginine signal peptide and suppresses signal peptide translocation activity such that transport via the Tat pathway is retarded. High-resolution, heteronuclear, multidimensional NMR spectroscopy reveals the 3D solution structure of NapD. The chaperone adopts a ferredoxin-type fold, which is completely distinct from the TorD family. Thus, NapD represents a new family of twin-arginine signal-peptide-binding proteins.
Organizational Affiliation:
Centre for Metalloprotein Spectroscopy and Biology, School of Biological Sciences, University of East Anglia, Norwich NR4 7TJ, United Kingdom.